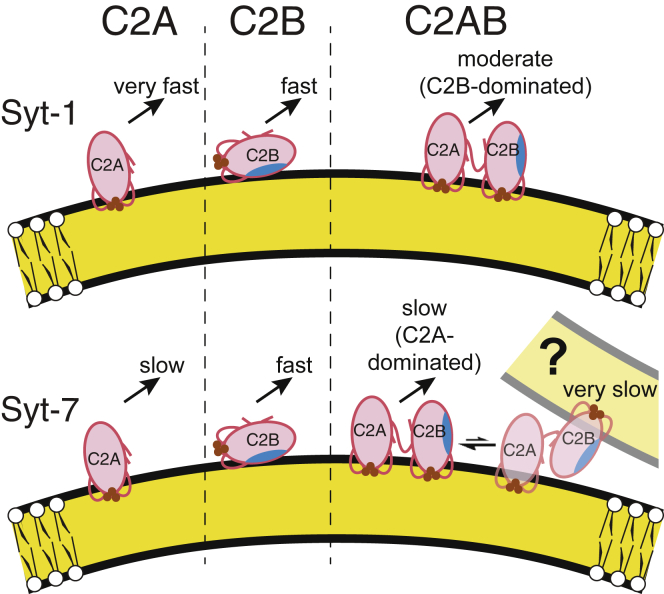
Figure 6.
Structural model of linkage effects on C2AB insertion and dissociation kinetics. (Left) The individual C2A domains of Syt-1 (top) and Syt-7 (bottom) penetrate membranes primarily via their CBLs (20, 21, 22, 23, 24) (Fig. 5, A and C); Syt-1 C2A dissociates much faster than Syt-7 C2A upon addition of EDTA (16) (Table 3). (Center) The individual C2B domains bind membranes relatively superficially through interaction of the polybasic region (blue shading) with lipid headgroups (30) along with modest insertion of CBL3 (23) (Fig. 5, E and G). (Right) In the C2AB tandem, linkage to C2A affects the binding geometry of the C2B domain, leading to deeper insertion of its CBL1 and CBL3 (23, 24) (Fig. 5, F–H). For Syt-1, this deeper insertion leads to much slower dissociation kinetics compared to the individual domains, i.e., interdomain cooperativity. For Syt-7, dissociation kinetics are dominated by the C2A domain and relatively unaffected by C2A-C2B linkage (Fig. 4). (Bottom right) A minor, very slow phase in Syt-7 C2AB dissociation kinetics may be explained by liposome clustering, which could include a subpopulation of C2AB fragments bound in trans to opposing liposomes. Brown circles represent Ca2+. Curvature is approximately to scale of 100-nm-diameter liposomes (Fig. S7). Kinetics upon EDTA addition are as follows: very fast, >500 s−1; fast, 80–200 s−1; moderate, 10–50 s−1; slow, 1–10 s−1; very slow, <1 s−1 (Table 3). To see this figure in color, go online.