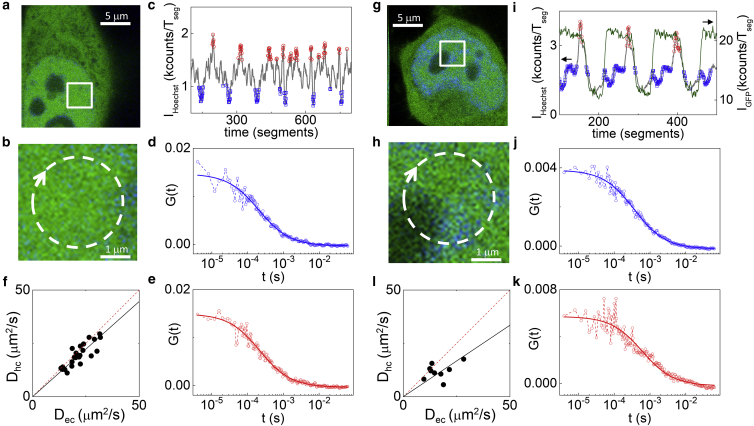
Figure 3.
Measurement of the diffusion coefficient of untagged GFP in eu- versus heterochromatin (a–f) and in euchromatin versus perinucleolar heterochromatin (g–k). (a and b) Each measurement is collected from a single nucleus stained with Hoechst (in blue in (a and b)), far from the nucleolus. Based on the Hoechst intensity trace (c), the FCS segments are sorted into two populations: the heterochromatin, in red, and the euchromatin, in blue. (d and e) Shown are sorted ACFs associated to the euchromatin (d) and the heterochromatin (e) along with the corresponding single component pure diffusion model fits ((d) D = 31.5 μm2/s; (e) D = 29.4 μm2/s). (f) Scatter plot of Dhc versus the value of Dec measured on the same cell is shown. Data represent measurements from different cells in a single experiment. The black solid line is a linear fit of the data with slope 0.89 ± 0.03. (g–i) For selecting the perinucleolar heterochromatin, the beams are scanned through the periphery of a nucleolus (g and h), and both the GFP (green line) and Hoechst (gray line) intensities are used as references (i). (j and k) Shown are sorted ACFs corresponding to the euchromatin (j) and perinucleolar heterochromatin (k), along with the corresponding single component pure diffusion model fits ((j) D = 17.6 μm2/s; (k) D = 10.6 μm2/s). (l) Scatter plot of Dhc versus Dec for perinuclear heterochromatin is shown. Data represent measurements from different cells in a single experiment. The solid black line is a linear fit of the data with slope 0.7 ± 0.1. To see this figure in color, go online.