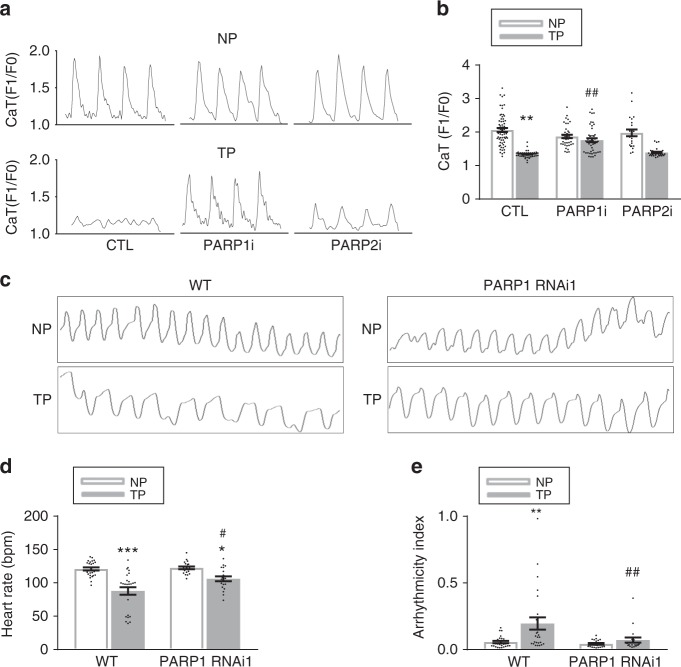
Fig. 3.
PARP1, not PARP2, is the key enzyme mediating tachypacing-induced contractile dysfunction in HL-1 cardiomyocytes and Drosophila. a, b Representative CaT traces and quantified CaT amplitude data in control nonpaced (NP) or tachypaced (TP) HL-1 cardiomyocytes transfected with scrambled siRNA (CTL), PARP1 siRNA (PARP1i), and PARP2 si RNA (PARP2i). **P < 0.01 vs. CTL NP, ##P < 0.01 vs. CTL TP. n = 62 cardiomyocytes for CTL NP, n = 39 for PARP1i NP, n = 22 for PARP2i NP, n = 56 for CTL TP, n = 47 for PARP1i TP, n = 27 for PARP2i TP. c Representative traces (10 s) prepared from high-speed movies of Drosophila prepupae. Movies were made from nonpaced (NP) and tachypaced (TP) Drosophila prepupae in wild-type (WT) and PARP1 knockdown (PARP1 RNAi1) strains. d, e Quantified heart rate (bpm: beats per minute) and arrhythmicity index in milliseconds (ms). Arrhythmicity index was defined as the standard deviation of the heart periodicity. *P < 0.05, **P < 0.01, ***P < 0.001 vs. WT NP, #P < 0.05 vs. WT TP, n = 26 Drosophila prepupae for WT, n = 20 Drosophila prepupae for PARP1i. Data are all expressed as mean ± s.e.m. Individual group mean differences were evaluated with the two-tailed Student’s t test