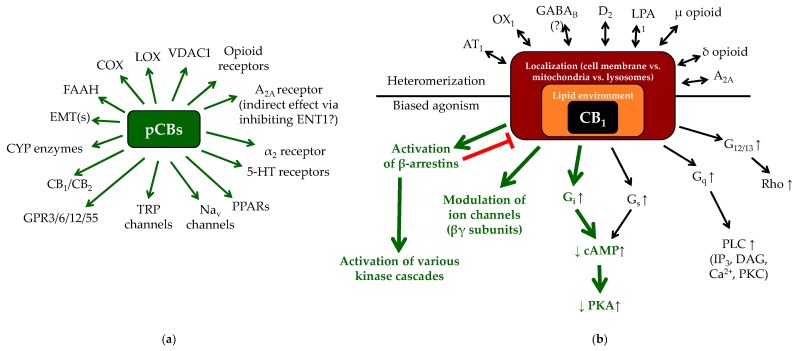
Figure 2.
Examples of the context-dependent complexity of the cannabinoid signaling. (a) Overview of the most important potential targets of the phytocannabinoids (pCBs), which can be concentration-dependently activated/antagonized/inhibited by these molecules. Each pCB can be characterized by a unique molecular fingerprint, and every pCB was found to interact with only a subset of potential targets shown on panel (a). Importantly, the interactions can even result in opposing outcomes (e.g., THC is a partial CB1 agonist, whereas CBD is a CB1 antagonist/inverse agonist), making prediction of cellular effects of the pCBs even more difficult. (b) The actual biological response, which develops following the activation of CB1 receptor depends on several additional factors, including biased agonism [31,32,65,66,67,68,69,70,71,72,73], possible receptor heteromerization [32,74,75,76,77,78,79,80], localization (i.e., cell membrane vs. mitochondria vs. lysosomes [81,82,83]), as well as the composition of the lipid microenvironment of the given membrane [58,84]. Green arrows on panel (b): the most common signaling pathways of CB1. Note that besides CB1, biased agonism is well-described in case of CB2, GPR18, GPR55 and GPR119 as well, whereas CB2 was proven to heteromerize with, e.g., C-X-C chemokine receptor type 4 chemokine receptor (CXCR4), or GPR55 (for details, see the above references). The question mark indicates that functional heteromerization of CB1 and GABAB receptors is questionable. AT1: angiotensin II receptor type 1; CYP: cytochrome P450 enzymes; D2: dopamine receptor 2; EMT(s): endocannabinoid membrane transporter(s); ENT1: equilibrative nucleoside transporter 1; GABAB: γ-aminobutyric acid receptor B; LPA1: lysophosphatidic acid receptor 1; Nav: voltage-gated Na+ channels; OX1: orexin 1 receptor; VDAC1: voltage-dependent anion channel 1. The figure was adapted and modified from [31] originally licensed under CC-BY, version 4.0.