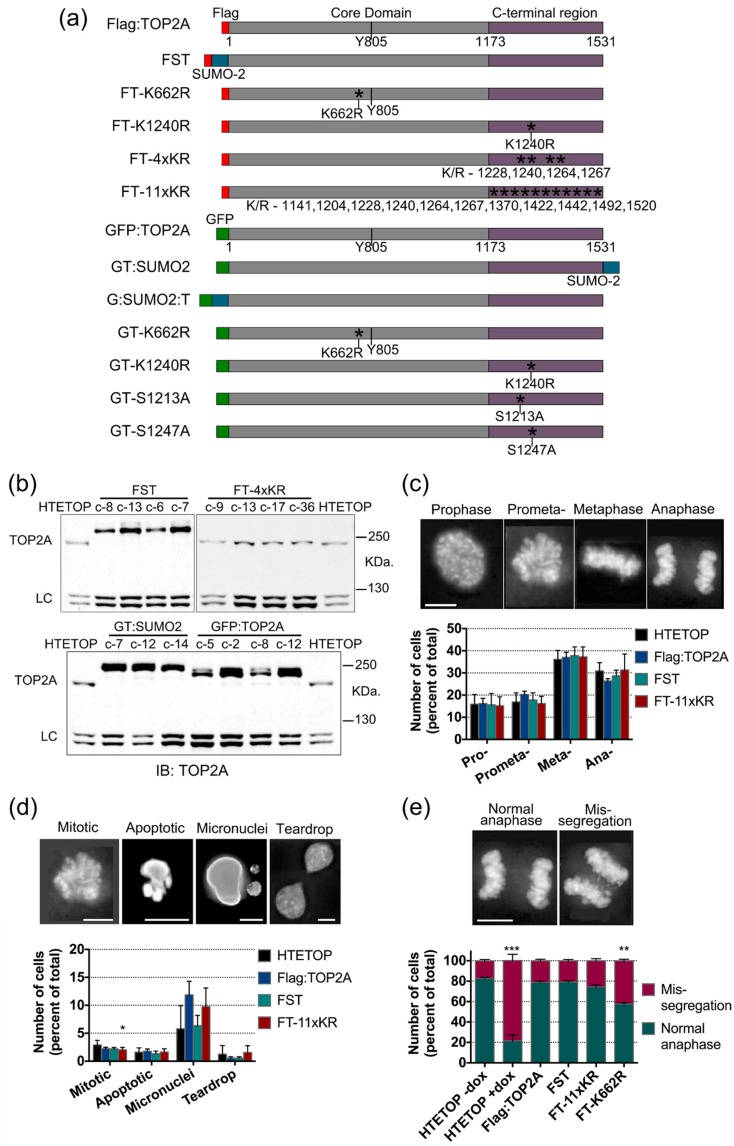
Figure 3.
Impact of TOP2A variants on mitotic progression and chromosome segregation. (a) Schematic showing the Flag- (red) and GFP (green)-tagged TOP2A variants studied in this work. (b) Representative immunoblots from HTETOP-derived transfectants stably expressing variant forms of TOP2A. Transfectants have been grown in the presence of doxycycline to repress the full length untagged TOP2A protein (~170 KDa) present in parental HTETOP cells. LC = an unidentified protein detected by the TOP2A antibody and described previously [45]. (c,d) Representative images and percentage of cells found at each stage of mitosis (c) and showing various nuclear phenotypes (d). Cells were scored by eye at 40× magnification. Three transfectant clones were tested for each variant and three independent experiments were performed. Approx. 500 mitotic cells and 1000 nuclei were scored per repeat. Slides were fixed in methanol and DNA stained with DAPI. Error bars represent standard error of the mean (s.e.m.). Significance was assessed by performing a one-way ANOVA analysis and post-hoc Tukey HSD test to compare lines to the HTETOP (-dox) control. (e) Representative images and percentage of cells, showing normal chromosome segregation and mis-segregation in anaphase. Cells were fixed in methanol and DNA stained with DAPI. Scale bar: 10 μm. Growth of the conditional null mutant line HTETOP in doxycycline for 48 h results in high levels of anaphase mis-segregation. Expressing Flag-tagged TOP2A in dox-treated HTETOP cells rescues the mis-segregation phenotype. Three transfectant clones were tested for each Flag-tagged variant. Three independent experiments performed with approx. 300 cells scored for each repeat. Error bars represent s.e.m. Significance was assessed by performing a one-way ANOVA to compare lines expressing the various Flag-tagged TOP2A variants to the HTETOP-dox control with the p value corrected using the post-hoc Tukey HSD analysis for multiple comparisons. *** p = 0.0001–0.001, ** p = 0.001–0.01, * p = 0.01–0.05.