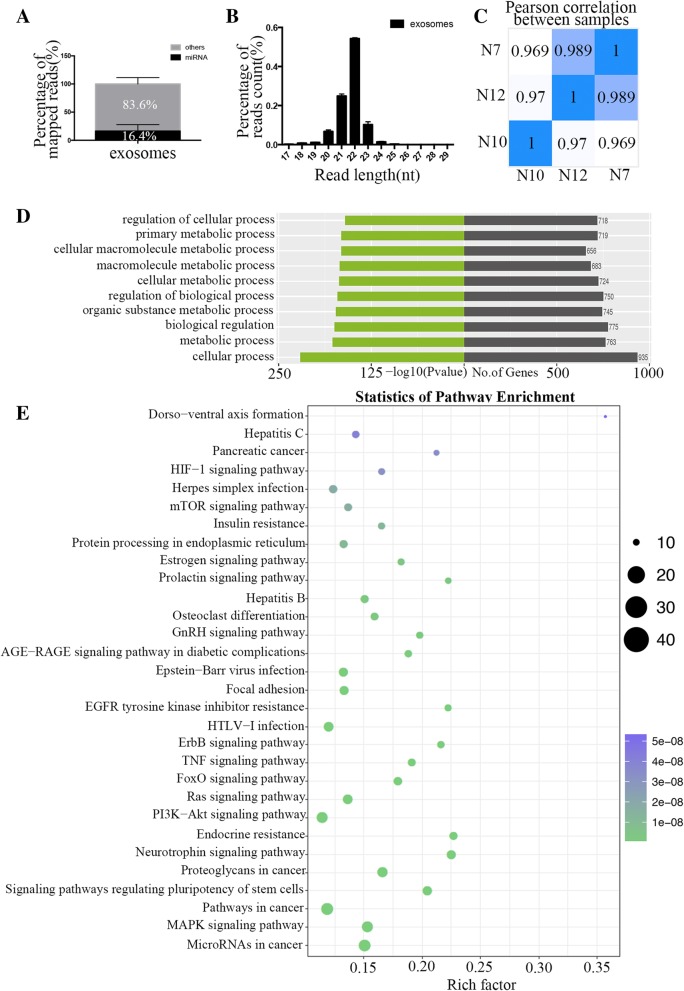
Fig. 4.
RNA-seq identified the miRNAs that are enriched in hADSC-derived exosomes and the predicted GO Terms and KEGG pathways that were targeted. a The average proportion of annotated miRNAs in the clean reads. b The size distribution of miRNAs reads was between 17 and 29 nt with a peak of 22 nt. c The Pearson correlation of three repeats was greater than 96.9%. The Gene ontology (GO) analysis (d) and Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis (e) of 1119 gene targets of the 30 most abundant miRNAs in the exosomes