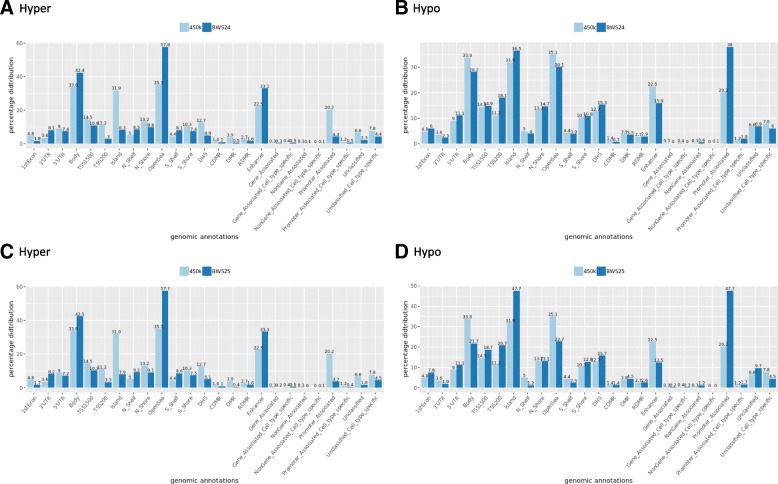
Fig. 2.
Genomic distribution of significant DMPs in patients BWS24 and BWS25. The plots compare the percentage of genomic distribution (genomic annotations of HumanMethylation450k array, Illumina) of all the informative probes included in our data and the percentage of genomic distribution of significant DMPs detected in patients BWS24 and BWS25. We compared the distribution of UCSC_RefGene_Group (1st exon, TSS200, TSS1500, 5′UTR, body, 3′UTR), Relation_to_UCSC_CpG_Island (Shore, Shelf, N, S), differentially methylated regions (DMR; CDMR, cancer-specific differentially methylated region; RDMR, reprogramming-specific differentially methylated region), overlapping enhancers loci, Regulatory_Feature_Group (Gene_Associated, Gene_Associated_Cell_type_specific, Promoter_Associated, Promoter_Associated_Cell_type_specific, Unclassified, Unclassified_Cell_type_specific), and DNase I hypersensitivity site (DHS). The number above each bar represents percentage. a The comparison of genomic distribution of hypermethylated DMPS in BWS24. b The comparison of genomic distribution of hypomethylated DMPs in BWS24. c The comparison of genomic distribution of hypermethylated DMPs in BWS25. d The comparison of hypomethylated DMPs in BWS25