Fig. 3 |. Preliminary analyses of CPD XR-seq data at a 1-h time point from treatment of human NHF1 cells with 10 J/m2 UV.
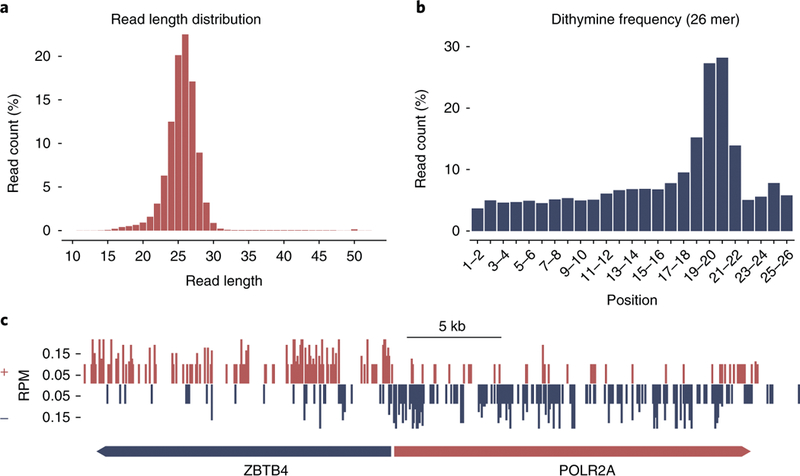
a, The read length distribution shows a single population (no degradation products) following TFIIH immunoprecipitation. b, Dithymine frequency is enriched at two main positions, 19–20 and 20–21, for the 26-nt excised oligomers obtained from CPD XR-seq. c, Screenshot of XR-seq signals in two genes, ZBTB4 and POLR2A, transcribed in opposite directions. Arrows indicate the direction of transcription. Repair asymmetry between strands that is due to transcription-coupled repair in the expressed genes is observed. The transcribed strands are the plus (above) and minus (below) strands for ZBTB4 and POLR2A, respectively. Therefore, those regions are repaired at higher levels because of transcription-coupled repair. The magnitude of the asymmetry varies depending on organism, gene and damage type. Data in c are from Hu et al.13. Histograms were plotted using the R software and the ggplot264 package, and the genomic region-specific repair levels were captured using IGV65.
