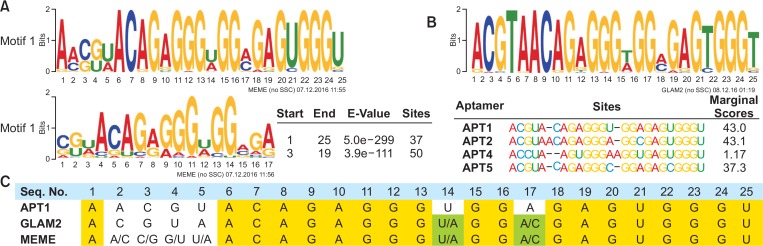
Fig. 3.
Consensus motifs analysis. (A, B) Consensus motifs in the random region were analyzed from 50 most popular aptamers of the S16 library using MEME (A) or GLAM2 (B) software. Sequence logos consist of stacks of letter for each position in the motif sequence. The height of the individual letter indicates the frequency of each nucleotide, and the positions where the motif starts were presented. Sites show the number of sequences contributing to the construction of the motif. The marginal scores of GLAM2 reflect the degree of matches between segments. (C) APT1 sequence was aligned with the most conserved motifs from MEME and GLAM2. Yellow color shows the sequences common in all motifs, whereas green color shows variable sequences in MEME and GLAM2 motifs.