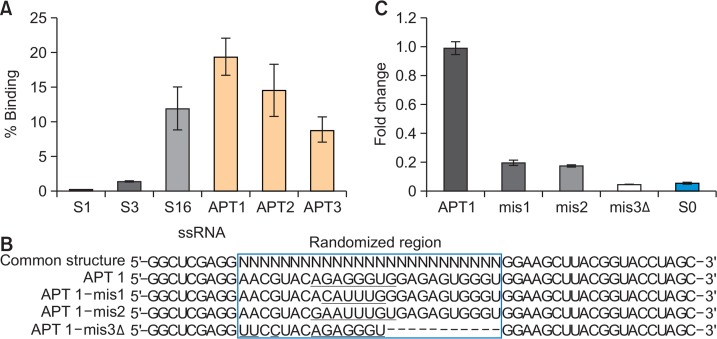
Fig. 5.
Evaluation of binding affinity of aptamers. (A) 1 μg of APT1, APT2, APT3, and three libraries were incubated with 1 μg of MLL1 protein and the binding activity was determined by qRT-PCR analysis. The percentage of bound RNA was expressed. (B) Mutation or deletion was introduced in APT1 as illustrated. The randomized region of APT1 are enclosed in a box, mutated nucleotides are underlined, and the deleted nucleotides are marked with hyphen. (C) The binding activity of each variant (1 μg) was measured by qRT-PCR analysis and the amount of bound RNA was calculated. Relative amount of bound RNAs was expressed as a fold change using wild-type APT1 was used as a control.