Despite much progress, we still do not have a clear understanding of how to elicit a protective neutralizing antibody response against HIV-1 through vaccination. There have been great strides in the development of envelope immunogens that mimic the virus particle, but less is known about how different vaccination modalities and adjuvants contribute to shaping the antibody response. We compared seven different vaccines that were administered to rhesus macaques and that delivered the same envelope protein through various modalities and with different adjuvants. The results demonstrate that some vaccine components are better than others at eliciting neutralizing antibodies with breadth.
KEYWORDS: human immunodeficiency virus, neutralizing antibodies, vaccines
ABSTRACT
The goals of preclinical HIV vaccine studies in nonhuman primates are to develop and test different approaches for their ability to generate protective immunity. Here, we compared the impact of 7 different vaccine modalities, all expressing the HIV-1 1086.C clade C envelope (Env), on (i) the magnitude and durability of antigen-specific serum antibody responses and (ii) autologous and heterologous neutralizing antibody capacity. These vaccination regimens included immunization with different combinations of DNA, modified vaccinia virus Ankara (MVA), soluble gp140 protein, and different adjuvants. Serum samples collected from 130 immunized monkeys at two key time points were analyzed using the TZM-bl cell assay: at 2 weeks after the final immunization (week 40/41) and on the day of challenge (week 58). Key initial findings were that inclusion of a gp140 protein boost had a significant impact on the magnitude and durability of Env-specific IgG antibodies, and addition of 3M-052 adjuvant was associated with better neutralizing activity against the SHIV1157ipd3N4 challenge virus and a heterologous HIV-1 CRF01 Env, CNE8. We measured neutralization against a panel of 12 tier 2 Envs using a newly described computational tool to quantify serum neutralization potency by factoring in the predetermined neutralization tier of each reference Env. This analysis revealed modest neutralization breadth, with DNA/MVA immunization followed by gp140 protein boosts in 3M-052 adjuvant producing the best scores. This study highlights that protein-containing regimens provide a solid foundation for the further development of novel adjuvants and inclusion of trimeric Env immunogens that could eventually elicit a higher level of neutralizing antibody breadth.
IMPORTANCE Despite much progress, we still do not have a clear understanding of how to elicit a protective neutralizing antibody response against HIV-1 through vaccination. There have been great strides in the development of envelope immunogens that mimic the virus particle, but less is known about how different vaccination modalities and adjuvants contribute to shaping the antibody response. We compared seven different vaccines that were administered to rhesus macaques and that delivered the same envelope protein through various modalities and with different adjuvants. The results demonstrate that some vaccine components are better than others at eliciting neutralizing antibodies with breadth.
INTRODUCTION
The goals of preclinical human immunodeficiency virus (HIV) vaccine studies in nonhuman primates (NHP) are to develop and test different approaches for their ability to generate protective immunity, understand how these responses are elicited and mediate protection, and translate the most efficacious approaches to human populations at risk for HIV-1 infection. NHP studies provide opportunities to investigate the impact of different types of vaccine vectors, immunogen forms, delivery routes, adjuvants, timing, and dosage by facilitating direct comparisons of variables, the synchronized collection of samples at optimal time points, and sampling more comprehensive than that which is possible in human subject cohorts (1, 2). NHP studies designed for cross-comparison can therefore lead to a better mechanistic understanding of protective immunity.
Heterologous vaccination strategies incorporating DNA prime and modified vaccinia virus Ankara (MVA) boost platforms have shown promise in preclinical and clinical testing of HIV vaccines (3). In humans, DNA/MVA approaches have exhibited excellent safety and immunogenicity profiles in phase I (HVTN-065) and phase IIa (HVTN-205) trials (4–6). Recombinant HIV-1 envelope (Env) protein gp120 immunizations have also exhibited robust safety and immunogenicity in two large phase III efficacy trials, VAX003 (7) and VAX004 (8). In addition, the more recently conducted RV144 phase III efficacy trial safely delivered a regimen consisting of a recombinant canarypox virus vector prime with bivalent Env gp120 protein boosts to over 8,000 individuals (9). This trial showed some evidence of reduced acquisition that was associated with antibody responses (10), although the interpretation of the statistical analysis remains under debate (11, 12). Nevertheless, follow-up studies (HVTN-100, HVTN-114, HVTN-702) have been undertaken to build upon the RV144 results by testing variables such as clade-matched Env immunogens, delayed protein and vector boosts, and different adjuvants. Thus, these established vaccination platforms that have been proven to be safe and effective in humans lend themselves to improvement by optimizing the Env immunogen, combining different vaccine modalities, and including novel adjuvants (13, 14).
To better understand how different vaccination modalities, immunogen forms, and adjuvants shape the immune response and influence the potential to elicit protective immunity, we previously performed a comparative analysis of the serum neutralizing antibody responses elicited in four DNA/MVA/protein-based simian immunodeficiency virus (SIV) vaccination challenge studies in rhesus macaques (trials M11, M12, M2, and M15) which together included 10 different regimens that expressed the SIVmac239 envelope (Env) glycoprotein (15–21). Gag/Pol immunogens, when included, were also derived from SIVmac239, and all trials used similar vaccine components and timelines, providing a unique opportunity to understand which components contributed to particular immune responses. For the comparative analysis, a panel of 14 genetically diverse SIVsm Envs consisting of tiers 1 to 3 was generated and used to assess neutralizing capacity in a manner similar to that used for neutralization of HIV-1 Envs (22, 23). Overall, a key finding of that study was that the different vaccination regimens predominantly elicited antibodies capable of neutralizing only the highly sensitive tier 1 SIVsm Envs (17). A conclusion from that study was that varying the antigen form, adjuvant, and delivery parameters did not alter the neutralizing antibody profile in the context of the SIVmac239 Env immunogen.
Subsequently, three additional vaccination trials in rhesus macaques that included seven different vaccination regimens were implemented utilizing the HIV-1 1086.C K160N Env immunogen instead of SIVmac239 (Fig. 1). The 1086.C Env was selected as the immunogen because it was derived from a clade C HIV-1-infected individual during acute infection (24). The immunogen Env sequence matched 12 of 25 Env sequences derived from the 1086 donor using single-genome PCR amplification (SGA) and was thus representative of the transmitted/founder (T/F) consensus sequence. This Env also displayed a desirable antigenic profile and exhibited a greater capacity to elicit antibodies with neutralization breadth in guinea pig immunization studies than a panel of other HIV-1 acute and chronic Env variants (25). Furthermore, the 1086.C Env gp120 protein was selected for inclusion in the bivalent subtype C gp120/MF59 protein component of the phase III HVTN-702 trial ongoing in South Africa (26). For the current study, all 1086.C Env immunogens encoded an N-linked glycan at position N160 (based on the strain HXB2 numbering), which is part of the V1V2 apex-directed, quaternary, broadly neutralizing antibody epitopes (27–29).
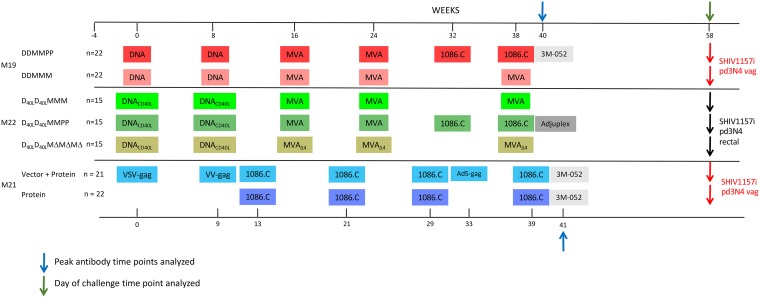
FIG 1.
Timeline of the vaccine studies in rhesus macaques. The immunization schedule for each vaccine trial (M19, M21, and M22) is shown on the timeline in weeks, with key time points provided. The vaccination arm of each trial and the number of animals included are indicated to the left. The agents used for immunization in each trial are indicated by colored boxes, highlighting similarities and differences between trials. Time points that were analyzed in this study are indicated by a blue arrow (peak antibody response at week 40 or 41, which is 2 weeks after the final immunization) or green arrow (time of SHIV challenge, week 58) above the timeline. Adjuvants that were used for the protein immunizations are indicated by gray boxes. Low-dose repeated mucosal challenges were carried out either intravaginally (M19, M21) or intrarectally (M22) and are described elsewhere.
Like the previous SIVmac239 Env-based studies, the HIV-1 Env immunogen trials M19, M21, and M22 also incorporated several novel approaches to enhance immunogenicity, with a particular focus on antibody responses. The M22 trial included the membrane-anchored rhesus macaque CD40 ligand (CD40L) adjuvant, which is coexpressed with Env on the surface of Gag-based virus-like particles produced by the DNA vaccine vector and which acts as a costimulatory molecule for dendritic cells and B cells (19). The CD40L adjuvant was previously associated with enhanced protection against SIVsmE660 challenge (19). One arm of the M22 trial also included an immunogenically enhanced version of the MVA vector lacking four immunomodulatory genes (interleukin-8 [IL-8] binding protein, soluble IL-1β receptor, CC chemokine binding protein, and a dominant negative Toll/IL-1 signaling adapter) (30). This MVA vector was previously shown to increase the magnitude of cellular and antibody immune responses against HIV-1 Env in rhesus macaques (30). Protein boost immunizations were included in one or more arms of the M19, M21, and M22 trials and consisted of the HIV-1 Env 1086.C K160N soluble gp140 protein formulated in one of two different adjuvants. The M22 trial utilized Adjuplex, which is composed of lecithin (submicron-sized liposomes) and a biodegradable carbomer homopolymer (31). Recent NHP studies of DNA/protein vaccination regimens demonstrated that Adjuplex is associated with promising neutralizing antibody titers and breadth (32, 33). The other adjuvant used in the M19 and M21 protein immunizations was a small molecule, 3M-052, encapsulated in poly(lactic-coglycoylic) acid (PLGA) nanoparticles. 3M-052 is a potent imidazoquinoline-based Toll-like receptor 7/8 (TLR7/8) agonist that results in prolonged retention of the adjuvant at the site of administration (34, 35). Finally, one vaccine arm in M21 also included three heterologous SIVmac239 Gag-expressing viral vectors (vesicular stomatitis virus [VSV] serotype New Jersey, the vaccinia virus [VV] WR strain, and adenovirus type 5 [Ad5]) to elicit robust Gag-directed CD8 T cell responses, in addition to anti-Env responses (C. Petitdemange, S. Kasturi, P. Kozlowski, R. Nabi, C. Quarnstrom, P. Reddy, C. Derdeyn, L. Spicer, P. Patel, T. Legere, Y. Kovalenkov, C. LaBranche, F. Villinger, M. Tomai, J. Vasilakos, B. Haynes, C. Kang, J. Gibbs, J. Yewdell, D. Barouch, J. Wrammert, D. Montefiori, E. Hunter, R. Amara, D. Masopust, and B. Pulendran, unpublished data).
In the present study, we assessed whether these vaccine modalities could impact the magnitude and durability of antigen-specific serum antibody responses and the autologous and heterologous neutralizing antibody capacity in the context of the widely used HIV-1 1086.C Env immunogen. Serum from two key time points was analyzed for 130 immunized monkeys: 2 weeks after the final immunization (week 40/41), which represents the peak serum antibody response, and the day of challenge (week 58), which occurred 17 to 18 weeks after the final immunization. The key findings are that inclusion of a gp140 protein boost, regardless of adjuvant, had a major positive impact on the magnitude and durability of Env-specific IgG antibodies. Protein boosts delivered in 3M-052 also produced better neutralizing activity against the simian-human immunodeficiency virus (SHIV) challenge virus and a heterologous HIV-1 Env, CNE8. To further investigate heterologous neutralization breadth, we utilized a well-characterized global panel of tier 2 Envs (23) and a newly described computational tool to objectively quantify neutralization potency (NP) by factoring in the predetermined sensitivity of the reference Envs (36). This analysis revealed that DNA/MVA immunization followed by gp140 protein boosts in 3M-052 adjuvant produced better serum neutralization breadth than the other vaccination regimens. Thus, in the context of HIV-1 Env 1086.C K160N, a combination of three different vaccine components, which each expressed a different form of Env, produced the broadest neutralization activity. The study highlights that protein-containing regimens, in particular, DNA/MVA/protein, provide a solid foundation for the development of novel adjuvants and trimeric Env immunogens that can eventually elicit a higher level of neutralizing antibody breadth.
RESULTS
Three preclinical HIV-1 clade C Env-based immunization studies with seven different regimens were carried out in rhesus macaques.
Three preclinical vaccination studies were carried out in rhesus macaques of Indian origin (Macaca mulatta), as depicted in Fig. 1. The M19 trial was comprised of two vaccination arms, both consisting of DNA immunizations at weeks 0 and 8 and two MVA boosts at weeks 16 and 24. After the second MVA immunization, the M19 groups received either 1086.C K160N Env gp140 protein in 3M-052 adjuvant at weeks 32 and 38 or an additional MVA immunization at week 38. Each vaccination group in M19 contained 22 animals. The M22 trial included three vaccination arms, all beginning with DNA immunizations at weeks 0 and 8 using a plasmid similar to that used in M19 but including rhesus macaque CD40 ligand (CD40L), which is incorporated into the virus particles produced. The MVA vaccines were administered at weeks 16, 24, and 38 with either the wild type or the MVAΔ4 deletion mutant, while a third arm received 1086.C K160N gp140 protein in Adjuplex adjuvant at weeks 32 and 38 in place of the last MVA vaccine. The vaccination groups in M22 contained either 14 or 15 monkeys. Finally, the M21 trial had two arms, neither of which included DNA or MVA vaccine vectors. Each group received 1086.C K160N gp140 protein immunizations in 3M-052 adjuvant at weeks 13, 21, 29, and 39; however, one arm also included immunization with three different heterologous viral vectors that expressed SIVmac239 Gag at weeks 0, 9, and 33. Each arm of the M21 trial contained 21 or 22 monkeys.
An intra-vaginal, low-dose repeated SHIV1157ipd3N4 challenge was carried out in M19 and M21 at week 58, which was 20 or 19 weeks after the final immunization, respectively. The results of the challenges are described in detail in two separate reports by V. Chamcha, P. Reddy, S. Kannanaganat, C. Wilkins, S. Gangadhara, V. Velu, R. Green, G. Law, J. Chang, J. Bowen, P. Kozlowski, M. Lifton, S. Santra, T. Legere, L. Chea, L. Chennareddi, M. Suthar, G. Silvestri, C. Derdeyn, M. Gale, Jr., F. Villinger, E. Hunter, and R. Amara (unpublished) and C. Petitdemange et al. (unpublished). For M22, an intrarectal, low-dose, repeated SHIV1157ipd3N4 challenge similar to that used in trials M19 and M21 was performed, and the results of the challenges are described in another report by T. Styles, S. Gangadhara, P. Reddy, S. Hicks, C. LaBranche, D. Montefiori, C. Derdeyn, P. Kozlowski, V. Velu, and R. Amara (unpublished). Thus, while these immunization regimens did not achieve the bar of protection, they did provide an opportunity to directly compare and contrast the responses elicited by distinct vaccine components and adjuvants. To do this, Env-specific IgG and neutralizing antibodies in serum were analyzed at two time points. Serum collected at week 40/41, 2 weeks after the final immunization, was chosen to represent the peak antibody response. Serum collected at week 58, the time of challenge, was also analyzed, as the response at this time point represents the serum antibody response closest to that encountered by the challenge virus.
Protein-containing vaccination regimens elicit higher-magnitude, more durable Env-specific IgG in serum.
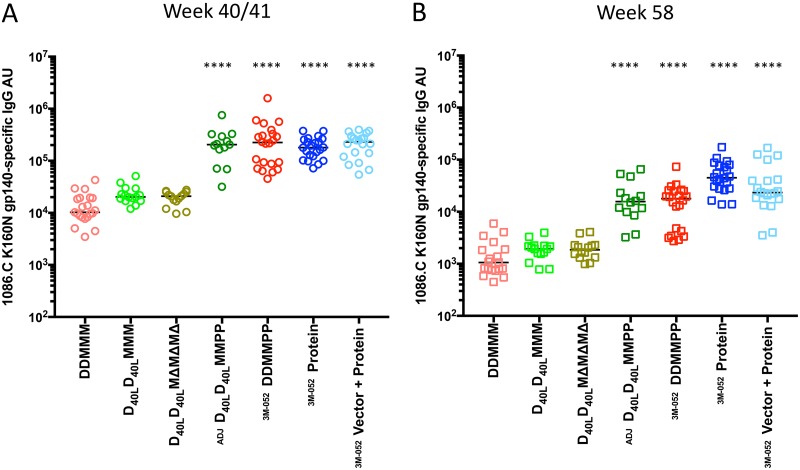
Figure 2 demonstrates the relative levels of 1086.C K160N Env gp140-specific IgG in serum detected by enzyme-linked immunosorbent assay (ELISA) across the seven different vaccination regimens. At the peak time point, which was week 40 for M19 and M22 and week 41 for M21, the median relative number of absorbance units (AU) for each group ranged from 10,263 for the group that received two DNA immunizations (weeks 0, 8) and three MVA immunizations (weeks 16, 24, 38) (DDMMM) to 230,286 for the vector-protein group (Fig. 2A). For the statistical analysis, the DDMMM vaccination group, which lacked protein and CD40L, and which utilized wild-type MVA, was used as a baseline comparator to test for differences in Env-specific serum IgG titers using a nonparametric Kruskal-Wallis test. This analysis revealed that the four regimens that included gp140 protein immunizations induced significantly higher Env-specific IgG levels than DDMMM (P < 0.0001 in all cases), while IgG levels in the non-protein-containing regimens with CD40L and the MVAΔ4 vector were not different from those in the DDMMM regimen. While a few animals in the two DNA/MVA/protein regimens had higher Env-specific IgG than animals in the protein-only regimens at week 40/41, the differences between the DNA/MVA/protein and protein-only groups were not significant.
FIG 2.
Immunogen-specific serum IgG levels induced by vaccination. The levels of IgG present in heat-inactivated, vaccinated monkey serum collected during the peak antibody response at week 40/41 (A) and on the day of challenge at week 54 or 58 (B) with the capacity to bind to the Env 1086.C K160N gp140 protein were quantified by ELISA. The relative absorbance units (AU) of Env-specific IgG was calculated for each sample by multiplying the OD450 of replicate wells by the serum dilution. The AU value is plotted on the y axis on a log10 scale. The vaccination modalities for each arm, as shown in Fig. 1, are indicated below the graph, organized by inclusion of protein boost. A horizontal bar indicates the median for each group. A Kruskal-Wallis test was performed to determine whether the levels of immunogen-specific serum IgG in each vaccination group were significantly different from those in the animals that received DDMMM, and significant differences are indicated by asterisks (****, P < 0.0001). The adjuvant used with the protein immunizations is indicated as Adjuplex (ADJ) or 3M-052. Week 54 samples were used instead of week 58 samples for 4 animals in the DDMMM, DDMMPP, protein, and vector-protein groups due to necropsy.
Overall, the protein-containing regimens produced, on average, 12-fold higher peak Env-specific IgG antibody levels than the DDMMM regimens. At week 58, the peak Env-specific IgG antibody levels in the four protein-containing regimens were again significantly higher than those in the DDMMM group (P < 0.0001), while those in the CD40L and the MVAΔ4 regimens were not different (Fig. 2B). However, the IgG levels for the DNA/MVA/protein regimens at week 58 had contracted more than those for the protein-only regimens. Overall, the protein-containing regimens at week 58 had Env-specific IgG levels that were 16-fold higher than those for the DDMMM regimens. The Env-specific IgG levels elicited by the protein-containing regimens declined by an average of 87% over the intervening 17- to 18-week period between the peak level and the level on the day of challenge, while the decline for the DNA/MVA regimens was slightly higher at an average of 90%. The protein-only regimen elicited the most stable Env-specific IgG levels, with IgG levels declining the least (75%) between the peak level and that on the day of challenge. In fact, using multiple pairwise comparisons at week 58, the level in the protein-only group was significantly higher than that in the group that received two MVA immunizations (weeks 16, 24) and two protein immunizations with HIV-1 1086.C K160N Env gp140 in Adjuplex vaccine adjuvant (D40LD40LMMPP) (P = 0.0027) and the group that received two DNA immunizations (weeks 0, 8), two MVA immunizations (weeks 16, 24), and two protein immunizations (weeks 32, 38) with HIV-1 Env 1086.C K160N gp140 in 3M-052 encapsulated in a PLGA-based nanoparticle adjuvant (DDMMPP) (P = 0.0001) but was not significantly different from that in the vector-protein group (P = not significant). Overall, these analyses demonstrate that the gp140 protein-containing regimens generated significantly higher peak Env-specific IgG levels that were more durable than the DNA/MVA-based regimens did. However, the decline in Env-specific IgG levels in all groups fell within the range for Env-specific serum IgG levels in rhesus macaques or human subjects vaccinated with similar regimens, which decreased from approximately 50% to 90% during the 12- to 20-week period following the final immunization and the peak antibody response in previous studies (5, 6, 16, 21, 37–39). Furthermore, the inclusion of CD40L in the DNA vaccine or the use of an immune-enhanced MVA vector did not have a positive or negative impact on the antigen-specific binding antibody responses in this context.
Low to moderate neutralizing activity against SHIV1157ipd3N4 Env was present at the peak but waned prior to challenge.
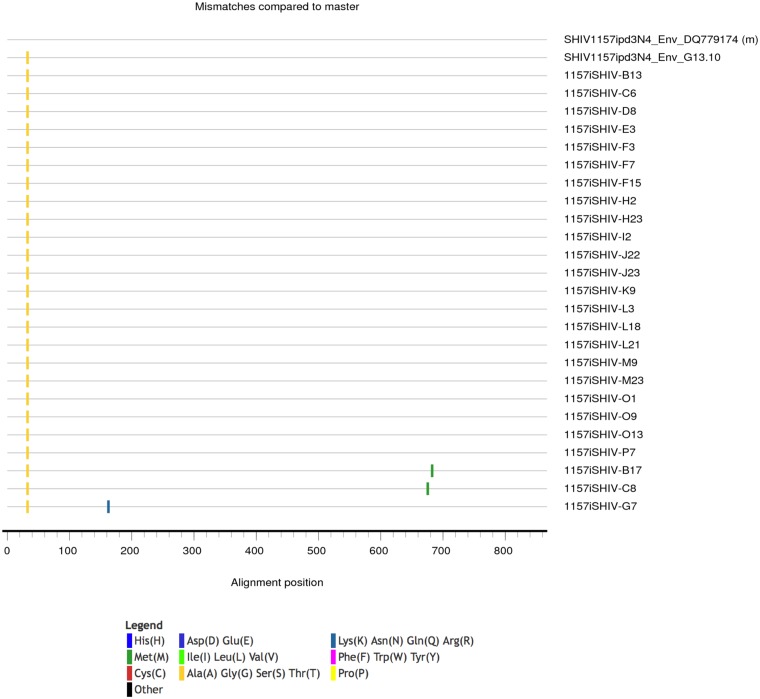
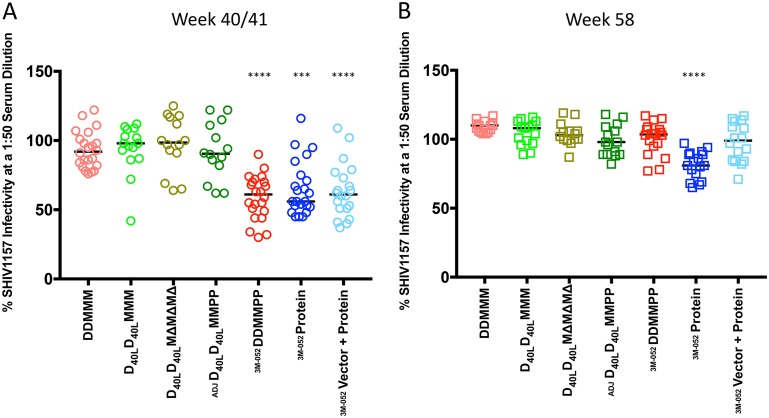
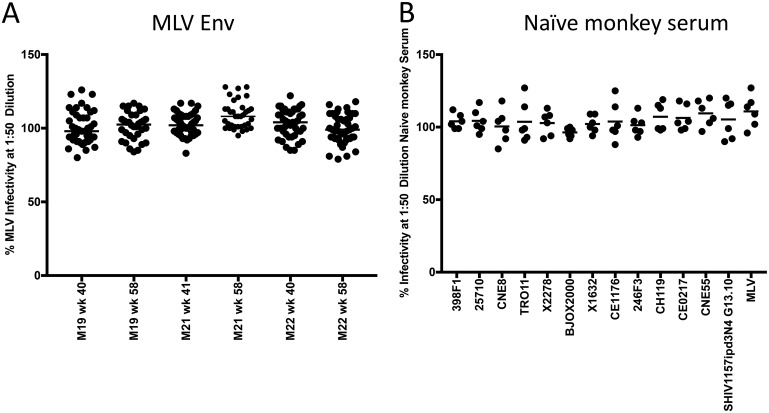
We next evaluated neutralization activity using an Env clone derived through single-genome PCR amplification (SGA) of the env gene from the clade C SHIV1157ipd3N4 heterologous challenge stock. This Env, designated SHIV1157ipd3N4 Env G13.10, was selected as its sequence is identical to the consensus sequence of the challenge stock and exhibited a single amino acid difference from the SHIV1157ipd3N4 sequence entry in GenBank (accession number DQ779174) that was present in all SGA-derived Env sequences (Fig. 3). Using a 1:50 dilution of week 40/41 serum from each monkey in the TZM-bl cell assay (Fig. 4), none of the serum samples were capable of complete neutralization; however, several did reduce infectivity to 50% or less. For comparison, the residual infectivity of murine leukemia virus (MLV) Env was above 79% for all vaccinated monkey serum samples, shown in aggregate in Fig. 5A, and naive monkey serum samples generated a very low background against all of the HIV-1 Envs tested, including SHIV1157 (Fig. 5B). The moderate levels of neutralization against SHIV1157ipd3N4 Env G13.10 were observed most consistently in 3 of the 4 protein-containing groups: the DDMMPP, protein, and vector-protein groups (Fig. 4A). These three groups were significantly different from the DDMMM group in their neutralization capacity (P < 0.001). Interestingly, the neutralization capacity of the other protein-containing regimen, D40LD40LMMPP, was not different from that of DDMMM, which indicated that the protein boost in this context did not elicit the same profile or magnitude of neutralization activity. This could be due to the delivery of the gp140 protein in Adjuplex instead of 3M-052 or other factors that have yet to be defined. Most of the neutralization activity against the challenge SHIV Env had waned by week 58, although interestingly, this activity in the protein-only immunization group appeared to be slightly greater, and this group was the only arm in which the activity was significantly different from that in the DDMMM arm (Fig. 4B). Overall, these findings demonstrate that the 1086.C K160N Env immunogens are capable of eliciting antibodies with neutralizing activity against SHIV1157ipd3N4; however, these antibodies lack potency and durability in serum, and it is unknown whether they were present at mucosal surfaces relevant to challenge.
FIG 3.
Highlighter plot of Env amino acid sequences derived from the SHIV1157pd3N4 challenge stock. A highlighter plot was generated (https://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter_top.html) using 25 SGA-derived sequences (Genbank accession numbers MK364746 through MK364771), the sequence of cloned Env G13.10, and the GenBank-derived Env sequence DQ779174 of the SHIV1157pd3N4 infectious molecular clone (as the master sequence). Ticks represent amino acid sequence differences from the master sequence and are defined in the key by color.
FIG 4.
Neutralization of a SHIV1157ipd3N4 Env pseudotype by vaccinated monkey serum. Dot plots were generated to show the residual percent infectivity of the SHIV1157ipd3N4 Env G13.10 by 1:50-diluted, heat-inactivated monkey serum collected at week 40/41 (A) and week 58 (B) arranged by vaccination group. The vaccination modalities for each arm are indicated below the graph. A horizontal bar indicates the median percent infectivity for each group. A Kruskal-Wallis test was performed to determine whether the levels of neutralization in each vaccination group were significantly different from those for the animals that received DDMMM, and significant differences are indicated by asterisks (***, P < 0.001; ****, P < 0.0001). The results for the protein-only groups that received 3M-052 adjuvant in trials M19 and M21 were significantly different from those for the DDMMM group. The adjuvant used with the protein immunizations is indicated as Adjuplex (ADJ) or 3M-052.
FIG 5.
Negative controls for neutralization using 1:50-diluted serum. (A) Residual infectivity of the MLV Env pseudovirus in the TZM-bl cell assay using all vaccinated monkey serum samples diluted 1:50, organized by trial (M19, M21, M22) and the time of sample collection (week 40/41 or 58), as indicated. The medians for all of the groups shown ranged from 98% to 108% residual infectivity. Of the 246 serum-Env combinations analyzed, 15 fell between 80% and 90% residual infectivity (6%) and 1 fell below 80% residual infectivity (0.4%). (B) Residual infectivity of a pool of naive rhesus macaque monkey serum diluted 1:50 against the global panel of reference Envs, the SHIV1137ipd3N4 Env, and the MLV Env. The monkey serum pool was included in each set of serum samples (see the heat maps in Fig. 8) according to the trial (M19, M21, M22) and the time of sample collection (weeks 40/41 and 58). Thus, the pool was run 6 times against each Env. The median residual infectivity for the HIV-1 Envs ranged from 96.5% to 109.5%. Of the 78 naive pooled serum–HIV-1 Env combinations analyzed, only 2 fell below 90% residual infectivity (2.5%).
Low to moderate neutralizing activity against heterologous tier 2 Envs from the global reference panel, but not the autologous Env, was present to varying degrees.
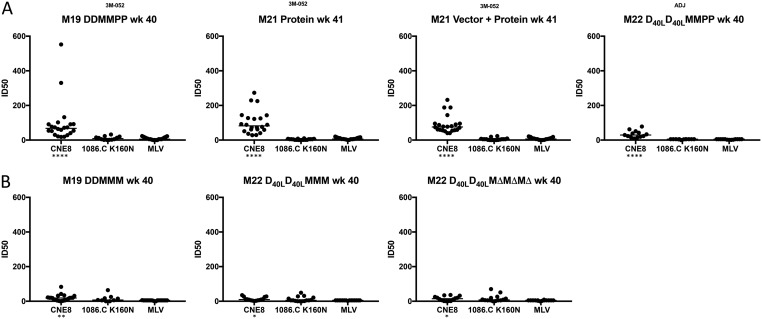
We next assessed the serum neutralization activity elicited by the different vaccination modalities against the autologous 1086.C K160N Env and a heterologous tier 2 Env, CNE8, in the TZM-bl cell assay. CNE8 is a CRF01_AE Env from the global reference panel that was isolated in China (Fig. 6) (23, 40). While CNE8 exhibits a tier 2 phenotype using HIV-positive serum samples in the TZM-bl cell assay, it is on the more sensitive side of the spectrum, providing an opportunity to detect antibody specificities different from those of the 1086.C K160N Env (23). Figure 6 shows that the serum neutralization activity against 1086.C K160N Env, presented as the 50% inhibitory dose (ID50) titer, was very low in all vaccination groups and was not significantly different from that against the MLV Env pseudovirus negative control. However, various levels of neutralization against CNE8 were observed. The individual ID50 titers calculated from the virus inhibition curves for CNE8 ranged from <1:20 to 1:552, and the median titers were significantly higher than those against MLV Env for all vaccination groups, determined using a Kruskal-Wallis test (P < 0.05). The highest titers against CNE8 were achieved in the four protein-containing regimens, with, again, higher responses being seen in the three groups that utilized the 3M-052 adjuvant, DDMMPP, protein, and vector-protein (Fig. 6A) (P < 0.0001). The DNA/MVA-based regimens also elicited CNE8-neutralizing activity that was above the background but that was at a lower level and less consistent than that in the protein-only groups (Fig. 6B).
FIG 6.
Determination of neutralization ID50 titer by vaccinated monkey serum against HIV-1 Env CNE8, HIV-1 Env 1086.C K160N, and MLV Env. Dot plots were generated to depict the neutralizing capacity of heat-inactivated, immunized monkey serum collected at week 40 for M19 and M22 and week 41 for M21. The TZM-bl cell assay was utilized to generate an infectivity curve in the presence of serially diluted serum, which was then used to calculate the 50% inhibitory dose (ID50) titer using Prism software. Each serum sample was evaluated in two independent assays with duplicate wells against HIV-1 Envs CNE8 and 1086.C K160N, while MLV Env was analyzed in a single assay with duplicate wells. The data are presented according to each vaccine arm, with the Env indicated below the graphs. The ID50 titer calculated in Prism (v.7) software is shown on the y axis for vaccination arms that included the 1086.C K160N protein boost (A) and the DNA/MVA-based vaccination arms without protein (B). A Kruskal-Wallis test was used to determine whether the neutralization of CNE8 or 1086.C K160N Env was significantly different from that for the negative control, MLV Env, for each panel, and significant P values are indicated below the graphs by asterisks (*, P < 0.05; **, P < 0.01; ****, P < 0.0001). The adjuvant used with the protein immunizations is indicated as Adjuplex (ADJ) or 3M-052.
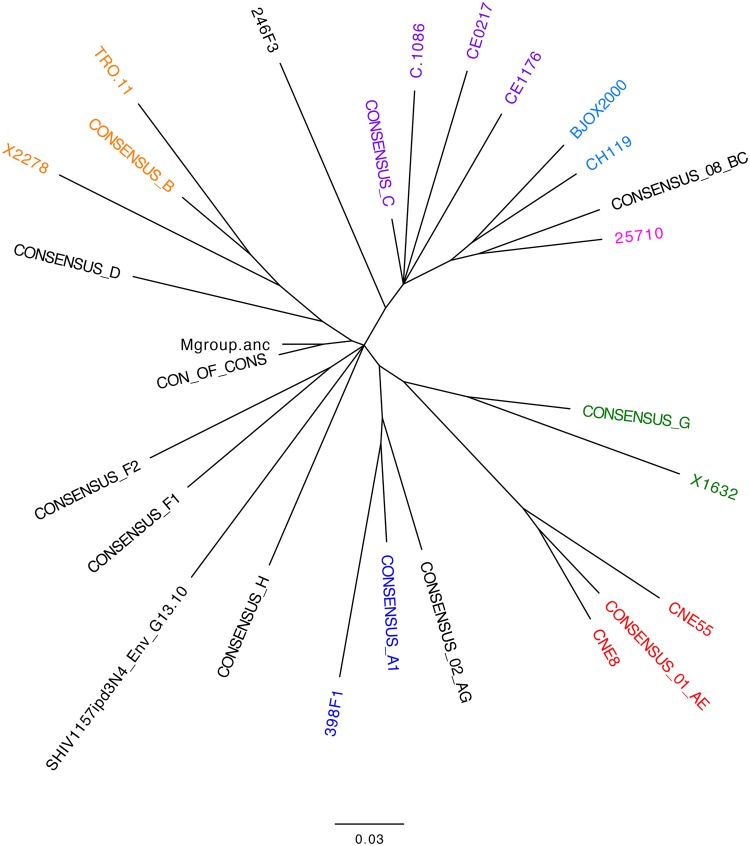
The consistent presence of moderate heterologous neutralizing activity against HIV-1 CNE8 prompted us to measure neutralization against the remaining 11 Envs in the global reference panel. Recently, a new computational tool was developed that assigns a quantitative potency score to serum samples, based on the predetermined tier of Envs that are neutralized, facilitating comparison across vaccination regimens (36). This tool, available on the Los Alamos HIV Database website at https://www.hiv.lanl.gov/content/sequence/NI/ni.html, accepts ID50 titers as well as binary data. We therefore utilized a single 1:50 dilution of serum to assess neutralization activity instead of a full dilution series, as complete neutralization and a reliable widespread calculation of ID50 titers were not expected. The threshold for neutralization was defined as less than 50% residual pseudovirus infectivity. The global panel was chosen to represent a range of diverse HIV-1 subtypes (Fig. 7) with a broad spectrum of neutralization sensitivities (Table 1) to provide the most informative outcomes (36).
FIG 7.
Phylogenetic tree of envelope variants used in the study. A neighbor-joining phylogenetic tree of Env amino acid sequences was generated for the 12 Env variants in the global reference panel, Env 1086.C, and SHIV1157ipd3N4 Env G13, using Geneious (v.9.0.4) software. The tree was exported and annotated in the FigTree (v.1.4.3) program. The horizontal bar indicates genetic distance. Clades are indicated by color: purple, clade C (African sequences); light blue, CRF07; green, clade G; red, CRF01; blue, clade A (subgroup 1); and orange, clade B. Mgroup.anc refers to the ancestral sequence for HIV-1 group M; CON_OF_CONS refers to the consensus of consensus sequences for subtypes A, B, C, D, F, G, and H. Both were generated using the Los Alamos National Laboratory HIV Database. Note that 25710 (pink) is a clade C sequence derived from India and 246F3 (black) is an HIV-1 clade A and C recombinant sequence.
TABLE 1.
Common names, neutralization indexes, GenBank accession numbers, and clades of HIV-1 global panel reference Envs
| Env | Common name | NIa | GenBank accession no. | Clade |
|---|---|---|---|---|
| 398_F1_F6_20 | 398F1 | 1.8 | HM215312 | A |
| 25710-2.43b | 25710 | 2.0 | EF117271 | C |
| CNE8 | CNE8 | 2.2 | HM215427 | CRF01 |
| BJOX002000.03.2 | BJOX2000 | 2.4 | HM215364 | CRF07 |
| X2278_C2_B6 | X2278 | 2.4 | FJ817366 | B |
| TRO.11 | TRO11 | 2.5 | AY835445 | B |
| X1632_S2_B10 | X1632 | 2.6 | FJ817370 | G |
| CE1176_A3 | CE1176 | 2.6 | FJ444437 | C |
| 246_F3_C10_2 | 246F3 | 2.6 | HM215279 | AC |
| CH119.10 | CH119 | 2.6 | EF117261 | CRF07 |
| CE703010217_B6 | CE0217 | 3.0 | FJ443575 | C |
| CNE55 | CNE55 | 3.0 | HM215418 | CRF01 |
NI, neutralization index. The neutralization index values were obtained from reference 36.
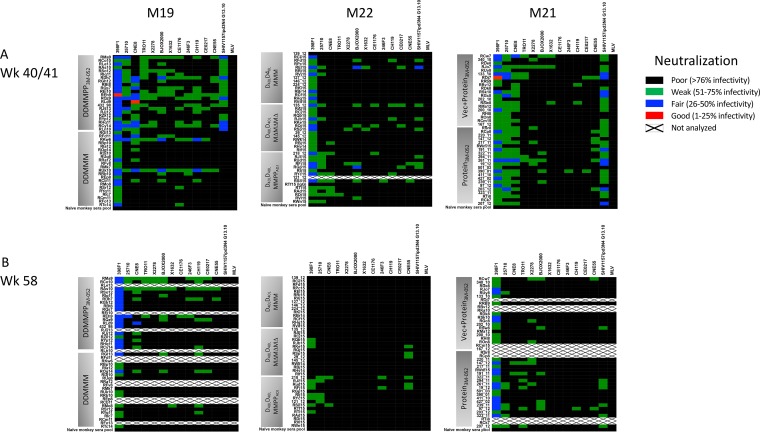
The neutralization heat maps displayed in Fig. 8A show that low but quantifiable levels of heterologous neutralization breadth were consistently detected against HIV-1 Envs 398 F1 (clade A), 25710 (clade C), and CNE8 (CRF01) in week 40/41 serum. These three Envs, while classified as tier 2, are on the more neutralization-sensitive side of the spectrum (23, 41). Less consistent neutralization of other, more resistant Envs in the panel was also observed at levels that were clearly distinguishable from those for the negative controls (MLV Env pseudovirus and naive monkey serum; also see Fig. 5). Overall, neutralizing antibody responses appeared to be the strongest in the three groups that received protein in 3M-052 adjuvant (M19 DDMMPP, M21 vector-protein, M21 protein), paralleling the neutralization of the SHIV Env and CNE8, which was also shown in this experiment. As expected, neutralization activity was stronger at week 40/41 than at week 58. We next set out to more objectively quantify the neutralization capacity of each serum sample, taking into account the tier or neutralization index (NI) of each Env (Table 1), in order to illuminate any potential differences between the vaccination regimens.
FIG 8.
Serum neutralization of the global HIV-1 Env reference panel. Heatmaps were generated to portray the neutralizing capacity of heat-inactivated immunized monkey serum from the peak response at week 40 for M19 and M22 and week 41 for M21 (A) and on the day of challenge at week 58 (B). A pool of naive monkey serum was included as a negative control against each Env (the last row in each heat map). Each heat-inactivated serum sample was diluted 1:50 and analyzed in duplicate against the HIV-1 Env pseudoviruses indicated at the top of each heat map using the TZM-bl cell assay. The Envs included the global reference panel, listed from the most to the least sensitive, as described previously (23) (n = 12); a representative Env cloned from the SHIV1157ipd3N4 challenge stock; and the MLV Env as a negative control. The trials are indicated at the top of the figure. For each heat map, the Envs are indicated at the top, and the vaccination arms are indicated to the left. The key presents the percent residual infectivity of each Env pseudovirus in the presence of each serum sample, represented as color. The color scale is the same for each heat map and indicates the potency of neutralization: red, good potency (1 to 25% residual infectivity); blue, fair potency (26 to 50% residual infectivity); green, weak potency (51 to 75% residual infectivity); and black, poor potency (greater than 76% residual infectivity). White boxes with an X, a serum-Env combination was not analyzed due to limited availability, necropsy, or poor sample quality. Vec, vector.
To briefly illustrate how the serum neutralization potency (NP) scores were calculated, representative plots generated at https://www.hiv.lanl.gov/content/sequence/NI/ni.html are shown in Fig. 9. Each individual serum sample was scored based on the Envs that it neutralized and the ranking on the neutralization index, shown on the x axis. All of the NP scores had well-supported P values for a nonzero slope (P < 0.1, chi-square likelihood-ratio test). The lowest NP score generated for our data set, 0.92, was calculated for serum samples that did not neutralize any of the 12 global reference Envs using the defined threshold (less than 50% residual infectivity at a 1:50 serum dilution) (Fig. 9A). The majority of serum samples at both time points fell into this category (Fig. 8). These serum samples would be expected to neutralize some heterologous tier 1 Envs but not tier 2 Envs. Neutralization of one of the more sensitive reference Envs, such as 25710 or 398F1, resulted in an NP score of 1.78 or 1.92 (Fig. 9B and C), which would indicate the capacity to neutralize most tier 1 Envs and a few tier 2 Envs. Neutralization of different combinations of sensitive and resistant Envs, such as 398F1, CNE8, BJOX2000 (BJOX), or CH119, resulted in NP scores of 2.18 and 2.23 (Fig. 9D and E), while neutralization of 398F1, 25710, and CNE8 produced an NP score of 2.32 (Fig. 9F). These serum samples could be expected to neutralize a subset of tier 2 Envs; however, no serum sample was able to neutralize an Env above a neutralization index of 2.6 (Table 1). Furthermore, none of the vaccine monkey serum samples analyzed here would be expected to have consistent and potent neutralization against tier 2 Envs. These NP scores are consistent with those for a panel of serum samples from rabbits immunized with various trimeric Env immunogens that was analyzed in a similar manner against a panel of tier 2 and 3 HIV-1 Envs determined using the Los Alamos online tool (36). Most of the serum samples in that study had no heterologous neutralization breadth and were assigned an NP score of 1.05; however, a subset (11 out of 50, 22%) had NP scores greater than 2, resulting from neutralization of a few tier 2 Envs, generally on the more sensitive side of the NI. For further comparison, we calculated serum NP scores using neutralization data from rhesus macaques immunized with various BG505 SOSIP.664 regimens, published in a previous study (42). The 9 vaccinated animals with the best antibody responses had a median NP score of 2.25 against the same global panel that was used here, with the NP scores ranging from 1.23 to 2.25. Those serum samples tended to show the best neutralization activity against 2 or 3 Envs with lower NIs. In our study, serum samples collected at week 40/41 from 17 immunized monkeys out of a total of 130 (13%) had NP scores of greater than 2, and this was also generally directed against Envs that fell closer to 2 than 3 on the NI scale.
FIG 9.
Representative serum neutralization potency graphs generated in the neutralization index tool. Each graph (A to F) shows the results of an analysis of 6 representative vaccinated monkey serum samples against the global reference panel of Envs. The Envs appear corresponding to their neutralization index, from 1 to 4, shown on the x axis. The calculated neutralization potency (NP) score indicates the virus tier that each serum can be expected to neutralize. NP is defined as the value for the fitted logistic function with an equal probability of neutralizing and not neutralizing a virus. Graphically, the NI value is where the fitted curve intersects 50% probability (equal odds of neutralizing and not neutralizing Envs tested). The P value gives the false-positive probability of rejecting the null hypothesis of no slope in the inferred logistic curve. NP scores in this study ranged from 0.92 (A), where none of the Envs were neutralized, to 2.32 (F), where 3 Envs at the lower side of the spectrum were neutralized. P values of <0.1 indicate a nonzero slope (chi-square likelihood-ratio test).
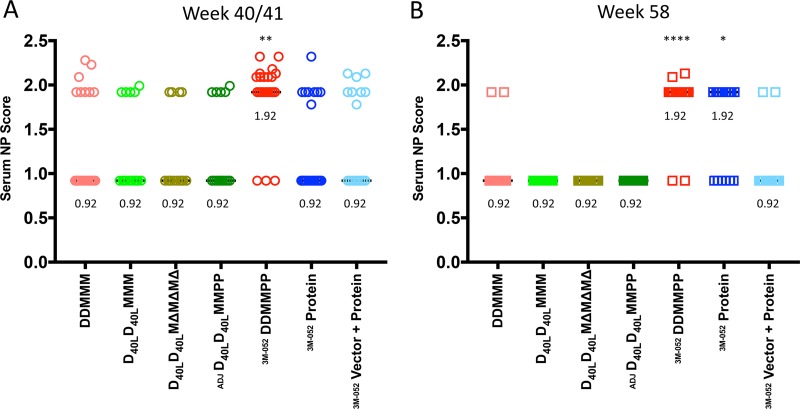
Using this computational approach, Fig. 10A demonstrates that the DDMMPP regimen at week 40 elicited the highest median NP score of 1.92. Only the score for the DDMMPP group was significantly different from that for the DDMMM group (P > 0.01). A substantial portion of the DDMMPP group serum samples (10 out of 22, 45%) achieved an NP score of 2.0 or greater. For comparison, only 3 DDMMM-immunized monkeys (13%) had NP scores of greater than 2.0. The remaining vaccination regimens elicited mainly tier 1 neutralizing capacity (median NP score, 0.92) at week 40/41, with a few exceptions. At week 58, the DDMMPP group also had the best heterologous neutralization capacity, followed by the protein-only group, with both groups having a median NP score of 1.92 (Fig. 10B). However, there were only two animals at this time point with NP scores greater than 2.0, and both were in the DDMMPP group. Thus, this approach revealed that the DDMMPP regimen generated a quantitatively greater and more frequent tier 2 neutralization capacity, albeit moderate, than the other regimens. A caveat to the study is that neutralization was measured at a single serum dilution, as opposed to a dilution series, which would have yielded a more precise quantitation of neutralization activity. Understanding the underlying mechanisms, including the contributions of the DNA and MVA immunizations, subsequent protein boosts, and adjuvant, could provide avenues for improvement, particularly by including stabilized Env trimers or rationally designed, epitope-based immunogens.
FIG 10.
Comparison of serum neutralization potency across vaccination regimens. The neutralization potency (NP) of each serum sample collected at week 40/41 (A) and week 58 (B) was calculated using the neutralization index tool from the Los Alamos HIV Database and is plotted on the y axis. The tool considers the neutralization susceptibility of each of the 12 reference Envs, as shown in Fig. 9. Each symbol represents the NP score of an individual monkey serum sample. An NP score of ∼1 indicates that a serum sample can be expected to neutralize only tier 1 Envs, while serum with increasing NP scores can be expected to neutralize more resistant tier 2 and 3 Envs. Most of the serum samples in this analysis had NP scores of 0.92 or 1.92, with a subset having NP scores exceeding 2. A Kruskal-Wallis test was used to determine whether the NP scores were significantly different from those found in the DDMMM-immunized animals. The DDMMPP immunization group was significantly different from the DDMMM group at both week 40 and week 58; the protein-only group was significantly different from the DDMMM group only at week 58 (*, P < 0.05; **, P < 0.01; ****, P < 0.0001). The adjuvant used with the protein immunizations is indicated as Adjuplex (ADJ) or 3M-052.
DISCUSSION
Numerous SIV/HIV vaccination platforms have been developed and tested in NHP for immunogenicity and protection, using different immunization routes, some of which were designed to elicit a particular type of immunity by systemic means or by targeting mucosal surfaces (1, 2). Here we compared several immunization approaches carried out under the Consortium for Non-Human Primate Models for AIDS Vaccine Research using a recently developed quantitative comparative tool to define neutralization breadth, in addition to standard comparative measurements. There is broad consensus in the field that inclusion of Env to induce an antibody component will be necessary to achieve high levels of protection. This is based in part on the observations that antibodies that neutralize in vitro using the standard TZM-bl cell assay have consistently shown strong, titer-dependent protection when passively administered to NHP (1). In contrast, passively administered nonneutralizing antibodies have not consistently mediated in vivo protection. Nevertheless, the only vaccination regimen associated with reduced HIV-1 acquisition in human subjects elicited antibodies that did not exhibit in vitro neutralizing capacity (10). Thus, it seems likely that multiple antibody effector functions can contribute to reduced acquisition, although it remains unclear how to purposefully direct the humoral immune response toward one or multiple effector functions. The RV144 trial also delivered Env in multiple forms, expressing Env proteins endogenously via a canarypox virus vector and as a bivalent soluble gp120 protein boost. Here, the best regimen for eliciting heterologous neutralizing capacity, DDMMPP, delivered the 1086.C K160N Env endogenously as gp160 (DNA) and as a cytoplasmic tail-truncated version, gp150 (MVA), on the surface of Gag-based particles and as soluble gp140. Understanding how the delivery of Env immunogens in different forms impacts antibody maturation and function or interferes with this process warrants further investigation.
A major goal of NHP vaccine studies is to identify strategies to elicit antibodies that can potently neutralize globally representative heterologous Env variants in vitro and then translate those approaches to humans. To date, there are relatively few examples of heterologous tier 2 neutralization breadth being raised in immunized rhesus macaques, and all of these successful studies have utilized optimized or purposefully selected Env immunogens. In one study, a DNA/gp140 coimmunization approach was used in rhesus macaques to deliver two strategically selected Env variants, which were each recovered from an HIV-1-infected individual at the onset of the development of neutralization breadth (33, 43). Using this strategy, heterologous neutralization activity against a small panel of tier 2 clade B and C Envs was observed using the A3R5 cell-based neutralization assay (44). Other studies have immunized rhesus macaques with stabilized, trimeric Env gp140 immunogens. In one case, a DNA plasmid vaccine encoding membrane-anchored, cleaved JR-FL Env, boosted by matching gp140-Foldon, which forms a stable trimer, elicited modest heterologous tier 2 neutralization activity detectable in the TZM-bl cell assay (32). The neutralization breadth was greater when assayed via the more sensitive A3R5 cell assay. In an additional study in which rhesus macaques were immunized with multiple BG505 SOSIP.664 trimer versions via numerous delivery routes, moderate neutralization breadth was observed in a subset of animals using the same global reference panel that we employed here (42). Although that study was focused primarily on autologous neutralization, heterologous neutralization breadth was observed primarily against reference Envs 398F1, 25710, CE1176, and X1632, which have NI values of 1.8, 2.0, 2.6, and 2.6, respectively. We also observed the most consistent neutralization activity against Envs 398F1 and 25710. A recent immunization study employed a rationally designed Env immunogen based on the HIV-1 Env gp41 fusion peptide that was administered to rhesus macaques in a regimen that included BG505 SOSIP.664 (45). Cross-clade neutralization breadth was observed against a panel of 58 HIV-1 variants that were matched for the vaccine’s fusion peptide amino acid sequence. Remarkably, serum from one of the five immunized rhesus macaques neutralized 41 of the 58 variants tested with variable ID50 titers, and the neutralization activity appeared to be directed at the fusion peptide. These studies collectively provide a proof of principle that levels of serum neutralization breadth can be generated using different Env immunogens and vaccination regimens, suggesting that it is important to pursue multiple vaccine approaches.
Our study, which generated weak tier 2 neutralization breadth that varied with the regimen, was based on the immunogenicity of the clade C Env 1086.C modified to contain the N160 glycan associated with V1V2/apex-directed broadly neutralizing antibodies (bnAbs) (46). Interest in using 1086.C Env as an immunogen began with an earlier study carried out in guinea pigs to identify promising HIV-1 Env immunogens in terms of inducing neutralizing antibody breadth (25). A goal of that study, which utilized gp140 immunogens, was to determine whether T/F Envs as a group had advantages over chronic or consensus Env variants in terms of eliciting neutralizing antibody breadth. Liao et al. found that T/F Envs tended to elicit antibodies that exhibited breadth and that could neutralize tier 1 and tier 2 Env variants (25). They also found that T/F Envs had a significantly higher binding affinity to monoclonal antibody (MAb) 17b, which recognizes a CD4-inducible (CD4i) epitope, even in the absence of soluble CD4 (sCD4). Env 1086.C was also recognized by MAbs A32, VRC01, and 19b and various V2 antibodies but was not bound by PG9 or PG16 due to the lack of N160. The authors assessed the strength of positive serum neutralization responses in immunized guinea pigs and found that 1086.C ranked as the second most effective immunogen. Because of its favorable antigenicity profile, it was included as one part of the bivalent clade C gp120 boost for the HVTN-100 phase I/II randomized, double-blind, clinical vaccine trial carried out in South Africa (47) and is also included in the subsequent ongoing phase IIb/III efficacy trial, HVTN-702 (26, 48). Overall, the subtype C bivalent gp120 immunogen has exhibited differences in antigenicity from the gp120s used in the RV144 regimen, in particular, for elicitation of responses directed at V1V2 (47). The 1086.C Env gp120 protein, when produced in CHO cells, also exhibited an unusually high occupancy of high-mannose glycans, which could favorably impact its antigenicity (26). Thus, while antigenicity cannot predict what type of antibody responses will be elicited by an Env immunogen, 1086.C possesses a number of desirable qualities supporting its broad use in antibody-based vaccination studies (49–51).
The 1086.C-based vaccine studies compared here elicited the most consistent neutralization activity against global reference Envs 398F1, CNE8, and, to a lesser extent, 25710 (Fig. 8). The NI for each of these Envs was 1.8, 2.2, and 2.0, respectively, putting them on the more susceptible side of the 1-to-4-NI spectrum (Table 1) (36) (see the neutralization index tool at https://www.hiv.lanl.gov/content/sequence/NI/ni.html). However, none of these Envs are particularly susceptible to neutralization by the CD4i MAb 17b, as discussed above, although 398F1 and CNE8 are susceptible to neutralization by high concentrations of this MAb in the absence of sCD4 (ID50 titers, 14 and 10 μg/ml, respectively) (23). Env 398F1 is also sensitive to a number of V3 MAbs and CD4 binding site (CD4bs) MAbs that typically have limited neutralization against other members of the global panel (23). Likewise, Env CNE8 is sensitive to gp41 cluster I-directed MAbs and a number of V1V2 MAbs with limited tier 2 neutralizing capacity (23). In contrast, Env 25710 does not exhibit any unusual sensitivity to nonneutralizing antibody specificities. Occasional neutralization of other Env variants, such as BJOX and CH119, with NIs of 2.4 and 2.6, respectively, was observed and could have also been mediated by recognition of poorly neutralizing epitopes in V3 or the CD4bs (23). Serum samples with peak antibody levels diluted 1:50 were capable of about 75% neutralization in the best cases, indicating that they could potentially reach complete neutralization in vitro at a lower dilution. Furthermore, ID50 titers assessed against Env CNE8 reached over 1:200 in a few cases. The observed neutralization breadth, while modest, could be directed at epitopes that are exposed in some Envs but not others and likely involves a polyclonal population, as opposed to a single neutralizing antibody specificity, such as the CD4bs. These studies therefore suggest that the 1086.C K160N Env, delivered through multiple vectored and protein boost strategies, does elicit some antibodies capable of neutralizing bona fide tier 2 variants at a moderate level in the TZM-bl cell assay.
This study provides new information about combinations of the Env immunogen, adjuvant, and delivery platform that could be used to elicit serum neutralization breadth. First, we consistently observed that including soluble gp140 protein boosts produces a higher magnitude of serum Env-specific IgG antibody responses with greater durability. All protein-containing regimens in our study, regardless of whether Adjuplex or 3M-052 adjuvant was used, elicited better serum Env-specific IgG responses than the DNA/MVA-based regimens. Second, groups with protein boosts were also associated with better neutralization of the challenge SHIV and low levels of heterologous neutralization breadth; however, this was limited to regimens that included the 3M-052 adjuvant, as the protein group that received Adjuplex was not different from the group that received DNA/MVA-based regimens in these contexts. Third, this study did not provide any evidence that inclusion of rhesus macaque CD40L in the DNA vaccine vector or the use of a modified MVA vector enhanced antibody responses in terms of magnitude, durability, or neutralizing activity. This finding is consistent with that of our previous study in the context of an SIV vaccine, where CD40L did not alter the serum neutralizing antibody profile (17); however, CD40L has been previously associated with enhanced protection against a mucosal SIV challenge and is worthy of further exploration and optimization (19). Furthermore, it is important to note that the CD40L-containing DNA/MVA/protein regimen cannot be directly compared to the DNA/MVA/protein regimen because two different adjuvants were used with the protein boosts. Fourth, neutralization of the autologous Env 1086.C K160N was poor in all vaccine groups. This Env has an NI of 2.4 and thus shares typical characteristics with Envs that are more resistant to neutralization. Finally, while moderate SHIV neutralization activity was present in some animals at week 40/41, these were low to undetectable in serum by the time of challenge at week 58. Protection generally requires serum antibody concentrations that are at least a log fold higher than the ID50 measured in vitro (1). These parallel comparisons of different vaccination regimens demonstrate that DDMMPP provides a promising foundation. This is consistent with the hypothesis that endogenous production of Env, Gag-Env particle formation, soluble protein boost, and adjuvant could all contribute in different ways to drive neutralizing antibody production. Furthermore, this vaccination platform has been used safely in human subjects to elicit antibody responses (4–6) and presents a strong foundation for improving neutralization breadth by strategies such as incorporating next-generation Env immunogens (52–55), extended vaccine delivery routes (42), and novel adjuvants (56). In conclusion, elicitation of modest serum neutralizing antibody responses against a subset of tier 2 HIV-1 Envs in NHP provides a step toward the goal of achieving protection against globally representative viral variants.
MATERIALS AND METHODS
Ethics statement.
The Emory University Institutional Animal Care and Use Committee (IACUC; AWA number A3180-01) approved these studies of nonhuman primates under protocols YER-2002540 and YER-2002761. These studies were performed in strict accordance with the United Stated Department of Agriculture regulations and the recommendations set forth in the Guide for the Care and Use of Laboratory Animals of the National Research Council (57). Animals were housed in pairs in standard nonhuman primate cages and received standard primate feed, as well as fresh fruit and enrichment daily. The animals had continuous and free access to water and enrichment resources, including objects for perching and other manipulanda. Animal welfare was monitored daily. Appropriate procedures were performed to ensure that potential distress, pain, or discomfort was alleviated. The sedative ketamine (5 to 10 mg/kg of body weight) or tiletamine and zolazepam (Telazol; 3 to 5 mg/kg) was administered before blood draws, which were performed by trained research staff.
Immunizations and samples.
Serum samples were obtained from a total of 130 Indian rhesus macaques (Macaca mulatta) immunized during three vaccine trials carried out at the Yerkes National Primate Research Center (designated M19 [n = 44], M21 [n = 43], and M22 [n = 45]; Fig. 1). M19 and M21 utilized only female monkeys, while each of the three groups in the M22 trial contained 10 males and 5 females. The M19 trial consisted of two vaccine groups. One group received two DNA immunizations (weeks 0, 8) and three MVA immunizations (weeks 16, 24, 38) (DDMMM), and the other group received two DNA immunizations (weeks 0, 8), two MVA immunizations (weeks 16, 24), and two protein immunizations (weeks 32, 38) with HIV-1 Env 1086.C K160N gp140 in 3M-052 encapsulated in a PLGA-based nanoparticle adjuvant (DDMMPP). The M21 trial consisted of two vaccine groups. One group received immunizations with heterologous recombinant viral vectors expressing full-length SIVmac239 Gag. These viral vectors were recombinant vesicular stomatitis virus (rVSV; serotype New Jersey) Gag (week 0) (58, 59), recombinant vaccinia virus (rVV; WR strain) Gag (week 9) (60, 61), and recombinant adenovirus type 5 (rAd5) Gag (week 37) (62), whichj were administered as four immunizations with HIV-1 Env 1086.C K160N gp140 in 3M-052 encapsulated in PLGA-based nanoparticle adjuvant (weeks 15, 21, 29, 39) (vector-protein); the second group received four immunizations of HIV-1 Env 1086.C K160N gp140 (weeks 15, 21, 29, 39) with no viral vectors (protein only). The M22 trial consisted of three vaccine groups. All three vaccine groups received two immunizations with DNA expressing rhesus macaque CD40 ligand (CD40L) (19) (weeks 0, 8). In addition, one group received MVA immunizations (weeks 16, 24, 38) (D40LD40LMMM); the second group received two MVA immunizations (weeks 16, 24) and two protein immunizations with HIV-1 1086.C K160N Env gp140 in Adjuplex vaccine adjuvant (catalog number A0362; Sigma) (weeks 32, 38) (D40LD40LMMPP); and the third group received three immunizations with an MVA deletion mutant (MVAΔ4) (30) (weeks 16, 24, 38) (D40LD40LMΔMΔMΔ). The DNA plasmid vector delivers the entire 1086.C K160N Env gp160 coding region along with the SIVmac239 Gag-, PR-, reverse transcriptase (RT)-, Tat-, and Rev-coding sequences. The MVA vector also encodes SIVmac239 Gag, RT, and PR but expresses the 1086.C K160N Env with the C-terminal 146 amino acids of the cytoplasmic tail truncated (gp150) to enhance stability and surface Env expression.
Immunization with a pGA-based DNA plasmid (20, 63) harboring SIVmac239 Gag/Pol and HIV-1 1086.C K160N Env gp160 (with or without the rhesus macaque CD40L-coding sequence) was carried out via intramuscular injection using a hypodermic needle at a 3-mg/dose; MVA expressing SIVmac239 Gag/Pol and HIV-1 1086.C K160N Env gp150 (64–66) was administered via intramuscular injection with 1 × 108 PFU/dose. Recombinant HIV-1 1086.C K160N Env gp140 protein was derived from an env gene cloned from an acutely infected clade C HIV-1-infected individual (24, 25). The cleavage site between gp120 and gp41 was mutated (R503E and R511E, HXB2 numbering), and the codon encoding lysine at position 160 was mutated to encode an asparagine, restoring the N-linked glycan. The protein was obtained from the Duke Human Vaccine Institute, where it was produced in 293F cells by transient transfection, purified by lectin chromatography, filtered using a 0.2-μm-pore-size filtration unit, and stored in phosphate-buffered saline (PBS; pH 7.4) at −80°C. Protein immunizations were also carried out subcutaneously and consisted of 100 μg/dose of 1086.C Env gp140 mixed with Adjuplex vaccine adjuvant in M22 (catalog number A0362; Sigma-Aldrich ) or the TLR7/8 agonist 3M-052 encapsulated in poly(lactic-coglycolic) acid (PLGA)-based nanoparticles in M19 and M21. The 3M-052 molecule was provided by 3M Drug Delivery Systems (St. Paul, MN, USA), poly(lactic-coglycolic) acid (PLGA; Mw, ∼12,000) polymer (Resomer RG502H) was procured from Boehringer Ingelheim (now supplied by Evonik Industries, AL), and poly(vinyl alcohol) (PVA; Mw, 31,000 to 50,000) was purchased from Sigma-Aldrich. 3M-052 encapsulated in PLGA nanoparticle formulations was synthesized using an oil-in-water (O/W) single emulsion, followed by a solvent evaporation process, previously described, with slight modifications (67). Nanoparticles were verified for size distribution using a dynamic light scattering (DLS)-based particle sizer (Brookhaven Instruments, NY). The encapsulation efficiency of 3M-052 was estimated using a UV-visible scan, with the peak being noted at 327 nm, as described before (9), and a standard curve was established with known amounts of 3M-052. rVV Gag and recombinant vaccinia virus (WR strain), each expressing full-length SIVmac239 gag, were generated as previously described (68–70). Heterologous viral vector immunizations included a 1-ml intravenous injection of 5 × 107 PFU of rVSV Gag at week 0. At 9 weeks, animals were boosted with a 1-ml intravenous injection of 1 × 107 PFU of rVV Gag. At 37 weeks, animals were then boosted with a 1-ml intravenous injection of 7.2 × 1010 viral particles of rAd5.
Neutralization assays.
Neutralization against HIV-1 Envs 1086.C K160N and CNE8 was measured using serially diluted, heat-inactivated immunized rhesus macaque serum in the TZM-bl cell assays as previously described, using cells that had been plated 1 day prior to the assay (15, 17, 30, 41, 71–80). In brief, Env pseudoviruses were generated by transfecting the Env-expressing plasmid DNA alongside the HIV-1 SG3ΔEnv proviral backbone DNA into 293T cells, collecting the supernatant 48 to 72 h later, clarifying it by centrifugation, and storing it in small aliquots at −80°C. Luciferase was quantified using a BioTek Synergy multimode microplate reader and Gen5 (v.3.02) software. Background luciferase activity was subtracted using the average for multiple control wells. The 50% inhibitory dose (ID50) titers were calculated from infectivity curves generated using Prism (v.7) software and the following method: log10 transformation of x values, normalization of y values from 0% to 100%, and nonlinear regression using a log(inhibitor)-versus-normalized response with variable slope function using a least-squares (ordinary) fit. Each assay was run with duplicate wells and repeated independently at least one time.
Neutralization was also measured against an SHIV1157ipd3N4 Env and a panel of 12 global reference HIV-1 Env clones (including CNE8) representative of the group M pandemic (23) using the same approach described above but with a higher throughput and a different format. A 1:50 dilution of each heat-inactivated serum sample was tested against each Env pseudovirus in duplicate wells, instead of using a dilution series. All assays were run in at least two independent assays, and the results were averaged. All neutralization results in the 1:50 dilution format are depicted in heat maps, generated in Prism (v.7) software, in Fig. 8. To quantitate the magnitude and breadth of neutralization, each serum-Env combination was assigned a binary score of 0 if residual viral infectivity was less than 50%, indicating sensitivity to neutralization; all others were assigned a score of 1, indicating resistance to neutralization. This assay was then used to calculate the neutralization potency score for each serum sample using the neutralization index tool at https://www.hiv.lanl.gov/content/sequence/NI/ni.html (36). Negative controls for serum neutralization activity included (i) a naive monkey serum pool and (ii) simian virus amphotropic murine leukemia virus (MLV) Env pseudovirus (Fig. 5).
1086.C K160N gp140-specific IgG ELISA.
Flat-bottom 96-well plates (catalog number 44-2404-21; Nunc MaxiSorp) were coated with 1 μg/ml of 1086.C K160N gp140 protein in carbonate-bicarbonate buffer (1.5 g Na2CO3, 2.9 g NaHCO3, 1 liter H2O) at 4°C overnight. The plates were then washed twice with PBS and blocked with 5% whey, 0.05% Tween in 1× PBS or 1% bovine serum albumin (BSA)–distilled H2O (dH2O) (10% BSA diluent/blocking solution; catalog number 50-61-10; KPL) for 2 h at room temperature. The plates were washed six times with PBS-Tween (diluted to 1× in dH2O; catalog number 50-63-00; KPL), and 100 μl of serum samples serially diluted in blocking buffer was added to the wells. The plates were incubated for 2 h at room temperature and then washed 6 times in PBS-Tween. Horseradish peroxidase-conjugated anti-monkey IgG (1 mg/ml; catalog number 70021; Alpha Diagnostic International) was diluted 1:10,000 in blocking buffer and added to each well, followed by incubation for 1 h at room temperature. The plates were washed 6 times in PBS-Tween, and then tetramethylbenzidine SureBlue 1-component microwell peroxidase substrate (catalog number 52-00-03; KPL) was added. The plate was covered with foil or placed in a drawer and incubated at room temperature for 15 min, at which time H2SO4 stop solution (4 N; catalog number SA818-1; Fisher Scientific) was added. The optical density at 450 nm (OD450) was determined by reading the absorbance on an Epoch microplate reader (BioTek), and the data were analyzed using Gen5 software. Quantitation of Env-specific serum IgG was performed by multiplying the absorbance reading at OD450 by the corresponding dilution factor and then calculating the average number of absorbance units (AU) from replicate wells for each sample.
Generation of an SHIV1157ipd3N4 Env clone.
An aliquot of the SHIV1157ipd3N4 challenge stock, which was obtained through the NIH AIDS Reference and Reagent Program (catalog number 11689) from Ruth Ruprecht, was used to isolate viral RNA using a Qiagen QIAamp viral RNA isolation kit (Germantown, MD) following the manufacturer’s instructions. cDNA was synthesized via RT-PCR using Superscript III reverse transcriptase (Thermo Fisher Scientific, Waltham, MA) and the reverse primer SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′) following the manufacturer’s instructions. Nested PCR was performed using procedures established in the Derdeyn lab for single-genome PCR amplification (SGA) (15, 18, 41, 73, 74, 76, 79, 81, 82). Briefly, the first round of PCR was performed in a 15-μl volume using Phusion HotStart II high-fidelity DNA polymerase (Thermo Scientific, Waltham, MA) and the following primers: SIVsm/macEnvF1 (5′-CCTCCCCCTCCAGGACTAGC-3′) and SIVsm/macEnvR1 (5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′). PCR was performed in a Bio-Rad C1000 Touch system with the following conditions: 94°C for 2 min; 35 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min; 68°C for 10 min; and a 4°C hold. The second round of PCR was performed in a 10-μl volume using Phusion HotStart II high-fidelity DNA polymerase (Thermo Scientific) and the following primers: SIVmacEnvF2 (5′-TATAATAGACATGGAGACACCCTTGAGGGAGC-3′) and SIVsmEnvR2 (5′-ATGAGACATRTCTATTGCCAATTTGTA-3′). PCR was performed in a Bio-Rad C1000 Touch system with the following conditions: 94°C for 2 min; 45 cycles of 94°C for 15 s, 55°C for 30 s, and 68°C for 4 min; 68°C for 10 min; and a 4°C hold. The PCR products were visualized on 0.8% agarose gels in 1× TAE (Tris-acetate-EDTA) and extracted for DNA purification using a Qiagen QIAquick gel extraction kit (Germantown, MD) following the manufacturer’s instructions. The nucleotide sequence was determined using the BigDye Terminator technology at Sequetech (Mountain View, CA), and contigs were assembled using the Sequencher (v.5.4.1) program (Gene Codes, Ann Arbor, MI). Confirmed sequences were imported into Geneious (v.9.0.4) software (Newark, NJ) to translate nucleotide sequences into amino acid sequences and create alignments. One PCR amplicon, G13.10, was deemed representative of the consensus sequence and was selected for directional one-step cloning into a cytomegalovirus-driven expression plasmid, pCDNA3.1V5HisTOPO/TA (catalog number K480040; Thermo Fisher), using established methods (41, 73, 74, 76, 79, 83).
ACKNOWLEDGMENTS
We are grateful to the veterinary staff and animal care technicians at the Yerkes National Primate Research Center for their assistance in these studies.
Funding was provided by the National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH): R01 AI-58706 and R01 AI-128837 (to C.A.D.), U19 AI-109633 (Integrated Preclinical/Clinical AIDS Vaccine Development Program to R.A.), and U19 AI-096187 (to R.A. and E.H.). Funding was also provided by Yerkes base grant ORIP/OD P51OD011132 (Yerkes National Primate Research Center).
The following reagents were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: panel of global HIV-1 Env clones (catalog number 12670) from David Montefiori (23), SHIV-1157ipd3N4 (catalog number 11689) from Ruth Ruprecht (84, 85), anti-HIV-1 gp120 monoclonal antibody 3BNC117 from Michel C. Nussenzweig (86), and the pSV-A-MLV-env plasmid (catalog number 1065) from Nathanial Landau and Dan Littman (87). The 1086.C Env plasmid was kindly provided by David Montefiori and used as a template to generate the 1086.C K160N mutant through site-directed mutagenesis. The soluble recombinant 1086.C K160N gp140 protein used in the immunizations and in the ELISAs was obtained from the Duke Human Vaccine Institute Protein Production Facility. It is also available from the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-1 gp140 recombinant protein (1086.C gp140C K160N) (catalog number 12580) from Barton F. Haynes and Hua-Xin Liao. (25, 88). We gratefully acknowledge Mark Tomai at 3M Drug Delivery Systems, St. Paul, MN, for the gift of 3M-052.
REFERENCES
- 1.Hessell AJ, Malherbe DC, Haigwood NL. 2018. Passive and active antibody studies in primates to inform HIV vaccines. Expert Rev Vaccines 17:127–144. doi: 10.1080/14760584.2018.1425619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sui Y, Gordon S, Franchini G, Berzofsky JA. 2013. Nonhuman primate models for HIV/AIDS vaccine development. Curr Protoc Immunol 102:14. doi: 10.1002/0471142735.im1214s102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chea LS, Amara RR. 2017. Immunogenicity and efficacy of DNA/MVA HIV vaccines in rhesus macaque models. Expert Rev Vaccines 16:973–985. doi: 10.1080/14760584.2017.1371594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goepfert PA, Elizaga ML, Sato A, Qin L, Cardinali M, Hay CM, Hural J, DeRosa SC, DeFawe OD, Tomaras GD, Montefiori DC, Xu Y, Lai L, Kalams SA, Baden LR, Frey SE, Blattner WA, Wyatt LS, Moss B, Robinson HL. 2011. Phase 1 safety and immunogenicity testing of DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis 203:610–619. doi: 10.1093/infdis/jiq105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goepfert PA, Elizaga ML, Seaton K, Tomaras GD, Montefiori DC, Sato A, Hural J, DeRosa SC, Kalams SA, McElrath MJ, Keefer MC, Baden LR, Lama JR, Sanchez J, Mulligan MJ, Buchbinder SP, Hammer SM, Koblin BA, Pensiero M, Butler C, Moss B, Robinson HL, HVTN 205 Study Group, National Institute of Allergy and Infectious Diseases HIV Vaccines Trials Network . 2014. Specificity and 6-month durability of immune responses induced by DNA and recombinant modified vaccinia Ankara vaccines expressing HIV-1 virus-like particles. J Infect Dis 210:99–110. doi: 10.1093/infdis/jiu003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchbinder SP, Grunenberg NA, Sanchez BJ, Seaton KE, Ferrari G, Moody MA, Frahm N, Montefiori DC, Hay CM, Goepfert PA, Baden LR, Robinson HL, Yu X, Gilbert PB, McElrath MJ, Huang Y, Tomaras GD, HIV Vaccines Trials Network (HVTN) 094 Study Group . 2017. Immunogenicity of a novel clade B HIV-1 vaccine combination: results of phase 1 randomized placebo controlled trial of an HIV-1 GM-CSF-expressing DNA prime with a modified vaccinia Ankara vaccine boost in healthy HIV-1 uninfected adults. PLoS One 12:e0179597. doi: 10.1371/journal.pone.0179597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pitisuttithum P, Gilbert P, Gurwith M, Heyward W, Martin M, van Griensven F, Hu D, Tappero JW, Choopanya K, Bangkok Vaccine Evaluation Group . 2006. Randomized, double-blind, placebo-controlled efficacy trial of a bivalent recombinant glycoprotein 120 HIV-1 vaccine among injection drug users in Bangkok, Thailand. J Infect Dis 194:1661–1671. doi: 10.1086/508748. [DOI] [PubMed] [Google Scholar]
- 8.Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF, rgp120 HIV Vaccine Study Group . 2005. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis 191:654–665. [DOI] [PubMed] [Google Scholar]
- 9.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH. 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 10.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gilbert PB, Berger JO, Stablein D, Becker S, Essex M, Hammer SM, Kim JH, Degruttola VG. 2011. Statistical interpretation of the RV144 HIV vaccine efficacy trial in Thailand: a case study for statistical issues in efficacy trials. J Infect Dis 203:969–975. doi: 10.1093/infdis/jiq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desrosiers RC. 2017. Protection against HIV acquisition in the RV144 trial. J Virol 91:e00905-17. doi: 10.1128/JVI.00905-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim JH, Excler JL, Michael NL. 2015. Lessons from the RV144 Thai phase III HIV-1 vaccine trial and the search for correlates of protection. Annu Rev Med 66:423–437. doi: 10.1146/annurev-med-052912-123749. [DOI] [PubMed] [Google Scholar]
- 14.Hsu DC, O'Connell RJ. 2017. Progress in HIV vaccine development. Hum Vaccin Immunother 13:1018–1030. doi: 10.1080/21645515.2016.1276138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burton SL, Kilgore KM, Smith SA, Reddy S, Hunter E, Robinson HL, Silvestri G, Amara RR, Derdeyn CA. 2015. Breakthrough of SIV strain smE660 challenge in SIV strain mac239-vaccinated rhesus macaques despite potent autologous neutralizing antibody responses. Proc Natl Acad Sci U S A 112:10780–10785. doi: 10.1073/pnas.1509731112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kasturi SP, Kozlowski PA, Nakaya HI, Burger MC, Russo P, Pham M, Kovalenkov Y, Silveira EL, Havenar-Daughton C, Burton SL, Kilgore KM, Johnson MJ, Nabi R, Legere T, Sher ZJ, Chen X, Amara RR, Hunter E, Bosinger SE, Spearman P, Crotty S, Villinger F, Derdeyn CA, Wrammert J, Pulendran B. 2017. Adjuvanting a simian immunodeficiency virus vaccine with Toll-like receptor ligands encapsulated in nanoparticles induces persistent antibody responses and enhanced protection in TRIM5alpha restrictive macaques. J Virol 91:e01844-16. doi: 10.1128/JVI.01844-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kilgore KM, Murphy MK, Burton SL, Wetzel KS, Smith SA, Xiao P, Reddy S, Francella N, Sodora DL, Silvestri G, Cole KS, Villinger F, Robinson JE, Pulendran B, Hunter E, Collman RG, Amara RR, Derdeyn CA. 2015. Characterization and implementation of a diverse simian immunodeficiency virus SIVsm envelope panel in the assessment of neutralizing antibody breadth elicited in rhesus macaques by multimodal vaccines expressing the SIVmac239 envelope. J Virol 89:8130–8151. doi: 10.1128/JVI.01221-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smith SA, Kilgore KM, Kasturi SP, Pulendran B, Hunter E, Amara RR, Derdeyn CA. 2016. Signatures in simian immunodeficiency virus SIVsmE660 envelope gp120 are associated with mucosal transmission but not vaccination breakthrough in rhesus macaques. J Virol 90:1880–1887. doi: 10.1128/JVI.02711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kwa S, Lai L, Gangadhara S, Siddiqui M, Pillai VB, Labranche C, Yu T, Moss B, Montefiori DC, Robinson HL, Kozlowski PA, Amara RR. 2014. CD40L-adjuvanted DNA/modified vaccinia virus Ankara simian immunodeficiency virus SIV239 vaccine enhances SIV-specific humoral and cellular immunity and improves protection against a heterologous SIVE660 mucosal challenge. J Virol 88:9579–9589. doi: 10.1128/JVI.00975-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai L, Kwa S, Kozlowski PA, Montefiori DC, Ferrari G, Johnson WE, Hirsch V, Villinger F, Chennareddi L, Earl PL, Moss B, Amara RR, Robinson HL. 2011. Prevention of infection by a granulocyte-macrophage colony-stimulating factor co-expressing DNA/modified vaccinia Ankara simian immunodeficiency virus vaccine. J Infect Dis 204:164–173. doi: 10.1093/infdis/jir199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lai L, Kwa SF, Kozlowski PA, Montefiori DC, Nolen TL, Hudgens MG, Johnson WE, Ferrari G, Hirsch VM, Felber BK, Pavlakis GN, Earl PL, Moss B, Amara RR, Robinson HL. 2012. SIVmac239 MVA vaccine with and without a DNA prime, similar prevention of infection by a repeated dose SIVsmE660 challenge despite different immune responses. Vaccine 30:1737–1745. doi: 10.1016/j.vaccine.2011.12.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.deCamp A, Hraber P, Bailer RT, Seaman MS, Ochsenbauer C, Kappes J, Gottardo R, Edlefsen P, Self S, Tang H, Greene K, Gao H, Daniell X, Sarzotti-Kelsoe M, Gorny MK, Zolla-Pazner S, LaBranche CC, Mascola JR, Korber BT, Montefiori DC. 2014. Global panel of HIV-1 Env reference strains for standardized assessments of vaccine-elicited neutralizing antibodies. J Virol 88:2489–2507. doi: 10.1128/JVI.02853-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abrahams MR, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, Ping LH, Athreya GS, Treurnicht FK, Keele BF, Wood N, Salazar-Gonzalez JF, Bhattacharya T, Chu H, Hoffman I, Galvin S, Mapanje C, Kazembe P, Thebus R, Fiscus S, Hide W, Cohen MS, Karim SA, Haynes BF, Shaw GM, Hahn BH, Korber BT, Swanstrom R, Williamson C. 2009. Quantitating the multiplicity of infection with human immunodeficiency virus type 1 subtype C reveals a non-Poisson distribution of transmitted variants. J Virol 83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao HX, Tsao CY, Alam SM, Muldoon M, Vandergrift N, Ma BJ, Lu X, Sutherland LL, Scearce RM, Bowman C, Parks R, Chen H, Blinn JH, Lapedes A, Watson S, Xia SM, Foulger A, Hahn BH, Shaw GM, Swanstrom R, Montefiori DC, Gao F, Haynes BF, Korber B. 2013. Antigenicity and immunogenicity of transmitted/founder, consensus and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol 87:4185–4201. doi: 10.1128/JVI.02297-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zambonelli C, Dey AK, Hilt S, Stephenson S, Go EP, Clark DF, Wininger M, Labranche C, Montefiori D, Liao HX, Swanstrom RI, Desaire H, Haynes BF, Carfi A, Barnett SW. 2016. Generation and characterization of a bivalent HIV-1 subtype C gp120 protein boost for proof-of-concept HIV vaccine efficacy trials in southern Africa. PLoS One 11:e0157391. doi: 10.1371/journal.pone.0157391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McLellan JS, Pancera M, Carrico C, Gorman J, Julien J-P, Khayat R, Louder R, Pejchal R, Sastry M, Dai K, O’Dell S, Patel N, Shahzad-Ul-Hussan S, Yang Y, Zhang B, Zhou T, Zhu J, Boyington JC, Chuang G-Y, Diwanji D, Georgiev I, Do Kwon Y, Lee D, Louder MK, Moquin S, Schmidt SD, Yang Z-Y, Bonsignori M, Crump JA, Kapiga SH, Sam NE, Haynes BF, Burton DR, Koff WC, Walker LM, Phogat S, Wyatt R, Orwenyo J, Wang L-X, Arthos J, Bewley CA, Mascola JR, Nabel GJ, Schief WR, Ward AB, Wilson IA, Kwong PD. 2011. Structure of HIV-1 gp120 V1/V2 domain with broadly neutralizing antibody PG9. Nature 480:336–343. doi: 10.1038/nature10696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pancera M, Shahzad-Ul-Hussan S, Doria-Rose NA, McLellan JS, Bailer RT, Dai K, Loesgen S, Louder MK, Staupe RP, Yang Y, Zhang B, Parks R, Eudailey J, Lloyd KE, Blinn J, Alam SM, Haynes BF, Amin MN, Wang LX, Burton DR, Koff WC, Nabel GJ, Mascola JR, Bewley CA, Kwong PD. 2013. Structural basis for diverse N-glycan recognition by HIV-1-neutralizing V1-V2-directed antibody PG16. Nat Struct Mol Biol 20:804–813. doi: 10.1038/nsmb.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker LM, Huber M, Doores KJ, Falkowska E, Pejchal R, Julien JP, Wang SK, Ramos A, Chan-Hui PY, Moyle M, Mitcham JL, Hammond PW, Olsen OA, Phung P, Fling S, Wong CH, Phogat S, Wrin T, Simek MD, Koff WC, Wilson IA, Burton DR, Poignard P. 2011. Broad neutralization coverage of HIV by multiple highly potent antibodies. Nature 477:466–470. doi: 10.1038/nature10373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garber DA, O'Mara LA, Gangadhara S, McQuoid M, Zhang X, Zheng R, Gill K, Verma M, Yu T, Johnson B, Li B, Derdeyn CA, Ibegbu C, Altman JD, Hunter E, Feinberg MB. 2012. Deletion of specific immune-modulatory genes from modified vaccinia virus Ankara-based HIV vaccines engenders improved immunogenicity in rhesus macaques. J Virol 86:12605–12615. doi: 10.1128/JVI.00246-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wegmann F, Moghaddam AE, Schiffner T, Gartlan KH, Powell TJ, Russell RA, Baart M, Carrow EW, Sattentau QJ. 2015. The carbomer-lecithin adjuvant Adjuplex has potent immunoactivating properties and elicits protective adaptive immunity against influenza virus challenge in mice. Clin Vaccine Immunol 22:1004–1012. doi: 10.1128/CVI.00736-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chakrabarti BK, Feng Y, Sharma SK, McKee K, Karlsson Hedestam GB, Labranche CC, Montefiori DC, Mascola JR, Wyatt RT. 2013. Robust neutralizing antibodies elicited by HIV-1 JRFL envelope glycoprotein trimers in nonhuman primates. J Virol 87:13239–13251. doi: 10.1128/JVI.01247-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malherbe DC, Pissani F, Sather DN, Guo B, Pandey S, Sutton WF, Stuart AB, Robins H, Park B, Krebs SJ, Schuman JT, Kalams S, Hessell AJ, Haigwood NL. 2014. Envelope variants circulating as initial neutralization breadth developed in two HIV-infected subjects stimulate multiclade neutralizing antibodies in rabbits. J Virol 88:12949–12967. doi: 10.1128/JVI.01812-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smirnov D, Schmidt JJ, Capecchi JT, Wightman PD. 2011. Vaccine adjuvant activity of 3M-052: an imidazoquinoline designed for local activity without systemic cytokine induction. Vaccine 29:5434–5442. doi: 10.1016/j.vaccine.2011.05.061. [DOI] [PubMed] [Google Scholar]
- 35.Van Hoeven N, Fox CB, Granger B, Evers T, Joshi SW, Nana GI, Evans SC, Lin S, Liang H, Liang L, Nakajima R, Felgner PL, Bowen RA, Marlenee N, Hartwig A, Baldwin SL, Coler RN, Tomai M, Elvecrog J, Reed SG, Carter D. 2017. A formulated TLR7/8 agonist is a flexible, highly potent and effective adjuvant for pandemic influenza vaccines. Sci Rep 7:46426. doi: 10.1038/srep46426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hraber P, Korber B, Wagh K, Montefiori D, Roederer M. 2018. A single, continuous metric to define tiered serum neutralization potency against HIV. Elife 7:e31805. doi: 10.7554/eLife.31805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kannanganat S, Wyatt LS, Gangadhara S, Chamcha V, Chea LS, Kozlowski PA, LaBranche CC, Chennareddi L, Lawson B, Reddy PB, Styles TM, Vanderford TH, Montefiori DC, Moss B, Robinson HL, Amara RR. 2016. High doses of GM-CSF inhibit antibody responses in rectal secretions and diminish modified vaccinia Ankara/simian immunodeficiency virus vaccine protection in TRIM5alpha-restrictive macaques. J Immunol 197:3586–3596. doi: 10.4049/jimmunol.1600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lai L, Vodros D, Kozlowski PA, Montefiori DC, Wilson RL, Akerstrom VL, Chennareddi L, Yu T, Kannanganat S, Ofielu L, Villinger F, Wyatt LS, Moss B, Amara RR, Robinson HL. 2007. GM-CSF DNA: an adjuvant for higher avidity IgG, rectal IgA, and increased protection against the acute phase of a SHIV-89.6P challenge by a DNA/MVA immunodeficiency virus vaccine. Virology 369:153–167. doi: 10.1016/j.virol.2007.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iyer SS, Gangadhara S, Victor B, Gomez R, Basu R, Hong JJ, Labranche C, Montefiori DC, Villinger F, Moss B, Amara RR. 2015. Codelivery of envelope protein in alum with MVA vaccine induces CXCR3-biased CXCR5+ and CXCR5− CD4 T cell responses in rhesus macaques. J Immunol 195:994–1005. doi: 10.4049/jimmunol.1500083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gnanakaran S, Daniels MG, Bhattacharya T, Lapedes AS, Sethi A, Li M, Tang H, Greene K, Gao H, Haynes BF, Cohen MS, Shaw GM, Seaman MS, Kumar A, Gao F, Montefiori DC, Korber B. 2010. Genetic signatures in the envelope glycoproteins of HIV-1 that associate with broadly neutralizing antibodies. PLoS Comput Biol 6:e1000955. doi: 10.1371/journal.pcbi.1000955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith SA, Burton SL, Kilembe W, Lakhi S, Karita E, Price M, Allen S, Hunter E, Derdeyn CA. 2016. Diversification in the HIV-1 envelope hyper-variable domains V2, V4, and V5 and higher probability of transmitted/founder envelope glycosylation favor the development of heterologous neutralization breadth. PLoS Pathog 12:e1005989. doi: 10.1371/journal.ppat.1005989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pauthner M, Havenar-Daughton C, Sok D, Nkolola JP, Bastidas R, Boopathy AV, Carnathan DG, Chandrashekar A, Cirelli KM, Cottrell CA, Eroshkin AM, Guenaga J, Kaushik K, Kulp DW, Liu J, McCoy LE, Oom AL, Ozorowski G, Post KW, Sharma SK, Steichen JM, de Taeye SW, Tokatlian T, Torrents de la Pena A, Butera ST, LaBranche CC, Montefiori DC, Silvestri G, Wilson IA, Irvine DJ, Sanders RW, Schief WR, Ward AB, Wyatt RT, Barouch DH, Crotty S, Burton DR. 2017. Elicitation of robust tier 2 neutralizing antibody responses in nonhuman primates by HIV envelope trimer immunization using optimized approaches. Immunity 46:1073–1088.e6. doi: 10.1016/j.immuni.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sather DN, Carbonetti S, Malherbe DC, Pissani F, Stuart AB, Hessell AJ, Gray MD, Mikell I, Kalams SA, Haigwood NL, Stamatatos L. 2014. Emergence of broadly neutralizing antibodies and viral coevolution in two subjects during the early stages of infection with human immunodeficiency virus type 1. J Virol 88:12968–12981. doi: 10.1128/JVI.01816-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hessell AJ, Malherbe DC, Pissani F, McBurney S, Krebs SJ, Gomes M, Pandey S, Sutton WF, Burwitz BJ, Gray M, Robins H, Park BS, Sacha JB, LaBranche CC, Fuller DH, Montefiori DC, Stamatatos L, Sather DN, Haigwood NL. 2016. Achieving potent autologous neutralizing antibody responses against tier 2 HIV-1 viruses by strategic selection of envelope immunogens. J Immunol 196:3064–3078. doi: 10.4049/jimmunol.1500527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu K, Acharya P, Kong R, Cheng C, Chuang G-Y, Liu K, Louder MK, O’Dell S, Rawi R, Sastry M, Shen C-H, Zhang B, Zhou T, Asokan M, Bailer RT, Chambers M, Chen X, Choi CW, Dandey VP, Doria-Rose NA, Druz A, Eng ET, Farney SK, Foulds KE, Geng H, Georgiev IS, Gorman J, Hill KR, Jafari AJ, Kwon YD, Lai Y-T, Lemmin T, McKee K, Ohr TY, Ou L, Peng D, Rowshan AP, Sheng Z, Todd J-P, Tsybovsky Y, Viox EG, Wang Y, Wei H, Yang Y, Zhou AF, Chen R, Yang L, Scorpio DG, McDermott AB, Shapiro L, Carragher B, Potter CS, Mascola JR, Kwong PD. 2018. Epitope-based vaccine design yields fusion peptide-directed antibodies that neutralize diverse strains of HIV-1. Nat Med 24:857–867. doi: 10.1038/s41591-018-0042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu X, Kong XP. 2016. Antigenic landscape of the HIV-1 envelope and new immunological concepts defined by HIV-1 broadly neutralizing antibodies. Curr Opin Immunol 42:56–64. doi: 10.1016/j.coi.2016.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bekker LG, Moodie Z, Grunenberg N, Laher F, Tomaras GD, Cohen KW, Allen M, Malahleha M, Mngadi K, Daniels B, Innes C, Bentley C, Frahm N, Morris DE, Morris L, Mkhize NN, Montefiori DC, Sarzotti-Kelsoe M, Grant S, Yu C, Mehra VL, Pensiero MN, Phogat S, DiazGranados CA, Barnett SW, Kanesa-Thasan N, Koutsoukos M, Michael NL, Robb ML, Kublin JG, Gilbert PB, Corey L, Gray GE, McElrath MJ, Team HP. 2018. Subtype C ALVAC-HIV and bivalent subtype C gp120/MF59 HIV-1 vaccine in low-risk, HIV-uninfected, South African adults: a phase 1/2 trial. Lancet HIV 5:e366–e378. doi: 10.1016/S2352-3018(18)30071-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang Z, Lorin C, Koutsoukos M, Franco D, Bayat B, Zhang Y, Carfi A, Barnett SW, Porter F. 2016. Comprehensive characterization of reference standard lots of HIV-1 subtype C gp120 proteins for clinical trials in southern African regions. Vaccines (Basel) 4:17. doi: 10.3390/vaccines4020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Eudailey JA, Dennis ML, Parker ME, Phillips BL, Huffman TN, Bay CP, Hudgens MG, Wiseman RW, Pollara JJ, Fouda GG, Ferrari G, Pickup DJ, Kozlowski PA, Van Rompay KKA, De Paris K, Permar SR. 2018. Maternal HIV-1 Env vaccination for systemic and breast milk immunity to prevent oral SHIV acquisition in infant macaques. mSphere 3:e00505-17. doi: 10.1128/mSphere.00505-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips B, Fouda GG, Eudailey J, Pollara J, Curtis AD II, Kunz E, Dennis M, Shen X, Bay C, Hudgens M, Pickup D, Alam SM, Ardeshir A, Kozlowski PA, Van Rompay KKA, Ferrari G, Moody MA, Permar S, De Paris K. 2017. Impact of poxvirus vector priming, protein coadministration, and vaccine intervals on HIV gp120 vaccine-elicited antibody magnitude and function in infant macaques. Clin Vaccine Immunol 24:e00231-17. doi: 10.1128/CVI.00231-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nelson CS, Pollara J, Kunz EL, Jeffries TL Jr, Duffy R, Beck C, Stamper L, Wang M, Shen X, Pickup DJ, Staats HF, Hudgens MG, Kepler TB, Montefiori DC, Moody MA, Tomaras GD, Liao HX, Haynes BF, Ferrari G, Fouda GGA, Permar SR. 2016. Combined HIV-1 envelope systemic and mucosal immunization of lactating rhesus monkeys induces a robust immunoglobulin A isotype B cell response in breast milk. J Virol 90:4951–4965. doi: 10.1128/JVI.00335-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward AB, Wilson IA. 2017. The HIV-1 envelope glycoprotein structure: nailing down a moving target. Immunol Rev 275:21–32. doi: 10.1111/imr.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bale S, Martine A, Wilson R, Behrens AJ, Le Fourn V, de Val N, Sharma SK, Tran K, Torres JL, Girod PA, Ward AB, Crispin M, Wyatt RT. 2018. Cleavage-independent HIV-1 trimers from CHO cell lines elicit robust autologous tier 2 neutralizing antibodies. Front Immunol 9:1116. doi: 10.3389/fimmu.2018.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kulp DW, Steichen JM, Pauthner M, Hu X, Schiffner T, Liguori A, Cottrell CA, Havenar-Daughton C, Ozorowski G, Georgeson E, Kalyuzhniy O, Willis JR, Kubitz M, Adachi Y, Reiss SM, Shin M, de Val N, Ward AB, Crotty S, Burton DR, Schief WR. 2017. Structure-based design of native-like HIV-1 envelope trimers to silence non-neutralizing epitopes and eliminate CD4 binding. Nat Commun 8:1655. doi: 10.1038/s41467-017-01549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanders RW, Moore JP. 2017. Native-like Env trimers as a platform for HIV-1 vaccine design. Immunol Rev 275:161–182. doi: 10.1111/imr.12481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ingale J, Stano A, Guenaga J, Sharma SK, Nemazee D, Zwick MB, Wyatt RT. 2016. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep 15:1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 58.Sacha JB, Chung C, Rakasz EG, Spencer SP, Jonas AK, Bean AT, Lee W, Burwitz BJ, Stephany JJ, Loffredo JT, Allison DB, Adnan S, Hoji A, Wilson NA, Friedrich TC, Lifson JD, Yang OO, Watkins DI. 2007. Gag-specific CD8+ T lymphocytes recognize infected cells before AIDS-virus integration and viral protein expression. J Immunol 178:2746–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Seder RA, Chang LJ, Enama ME, Zephir KL, Sarwar UN, Gordon IJ, Holman LA, James ER, Billingsley PF, Gunasekera A, Richman A, Chakravarty S, Manoj A, Velmurugan S, Li M, Ruben AJ, Li T, Eappen AG, Stafford RE, Plummer SH, Hendel CS, Novik L, Costner PJ, Mendoza FH, Saunders JG, Nason MC, Richardson JH, Murphy J, Davidson SA, Richie TL, Sedegah M, Sutamihardja A, Fahle GA, Lyke KE, Laurens MB, Roederer M, Tewari K, Epstein JE, Sim BK, Ledgerwood JE, Graham BS, Hoffman SL, VRC 312 Study Team . 2013. Protection against malaria by intravenous immunization with a nonreplicating sporozoite vaccine. Science 341:1359–1365. doi: 10.1126/science.1241800. [DOI] [PubMed] [Google Scholar]
- 60.Schwartz S, Campbell M, Nasioulas G, Harrison J, Felber BK, Pavlakis GN. 1992. Mutational inactivation of an inhibitory sequence in human immunodeficiency virus type 1 results in Rev-independent Gag expression. J Virol 66:7176–7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rosati M, von Gegerfelt A, Roth P, Alicea C, Valentin A, Robert-Guroff M, Venzon D, Montefiori DC, Markham P, Felber BK, Pavlakis GN. 2005. DNA vaccines expressing different forms of simian immunodeficiency virus antigens decrease viremia upon SIVmac251 challenge. J Virol 79:8480–8492. doi: 10.1128/JVI.79.13.8480-8492.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abbink P, Lemckert AA, Ewald BA, Lynch DM, Denholtz M, Smits S, Holterman L, Damen I, Vogels R, Thorner AR, O'Brien KL, Carville A, Mansfield KG, Goudsmit J, Havenga MJ, Barouch DH. 2007. Comparative seroprevalence and immunogenicity of six rare serotype recombinant adenovirus vaccine vectors from subgroups B and D. J Virol 81:4654–4663. doi: 10.1128/JVI.02696-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Smith JM, Amara RR, Campbell D, Xu Y, Patel M, Sharma S, Butera ST, Ellenberger DL, Yi H, Chennareddi L, Herndon JG, Wyatt LS, Montefiori D, Moss B, McClure HM, Robinson HL. 2004. DNA/MVA vaccine for HIV type 1: effects of codon-optimization and the expression of aggregates or virus-like particles on the immunogenicity of the DNA prime. AIDS Res Hum Retroviruses 20:1335–1347. doi: 10.1089/aid.2004.20.1335. [DOI] [PubMed] [Google Scholar]
- 64.Amara RR, Villinger F, Altman JD, Lydy SL, O'Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma HL, Grimm BD, Hulsey ML, Miller J, McClure HM, McNicholl JM, Moss B, Robinson HL. 2001. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Science 292:69–74. doi: 10.1126/science.1058915. [DOI] [PubMed] [Google Scholar]
- 65.Amara RR, Villinger F, Altman JD, Lydy SL, O’Neil SP, Staprans SI, Montefiori DC, Xu Y, Herndon JG, Wyatt LS, Candido MA, Kozyr NL, Earl PL, Smith JM, Ma H-L, Grimm BD, Hulsey ML, McClure HM, McNicholl JM, Moss B, Robinson HL. 2002. Control of a mucosal challenge and prevention of AIDS by a multiprotein DNA/MVA vaccine. Vaccine 20:1949–1955. doi: 10.1016/S0264-410X(02)00076-2. [DOI] [PubMed] [Google Scholar]
- 66.Van Rompay KK, Greenier JL, Cole KS, Earl P, Moss B, Steckbeck JD, Pahar B, Rourke T, Montelaro RC, Canfield DR, Tarara RP, Miller C, McChesney MB, Marthas ML. 2003. Immunization of newborn rhesus macaques with simian immunodeficiency virus (SIV) vaccines prolongs survival after oral challenge with virulent SIVmac251. J Virol 77:179–190. doi: 10.1128/JVI.77.1.179-190.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kasturi SP, Skountzou I, Albrecht RA, Koutsonanos D, Hua T, Nakaya HI, Ravindran R, Stewart S, Alam M, Kwissa M, Villinger F, Murthy N, Steel J, Jacob J, Hogan RJ, Garcia-Sastre A, Compans R, Pulendran B. 2011. Programming the magnitude and persistence of antibody responses with innate immunity. Nature 470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wu K, Kim GN, Kang CY. 2009. Expression and processing of human immunodeficiency virus type 1 gp160 using the vesicular stomatitis virus New Jersey serotype vector system. J Gen Virol 90:1135–1140. doi: 10.1099/vir.0.009019-0. [DOI] [PubMed] [Google Scholar]
- 69.An HY, Kim GN, Wu K, Kang CY. 2013. Genetically modified VSV(NJ) vector is capable of accommodating a large foreign gene insert and allows high level gene expression. Virus Res 171:168–177. doi: 10.1016/j.virusres.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 70.Bennink JR, Yewdell JW, Smith GL, Moller C, Moss B. 1984. Recombinant vaccinia virus primes and stimulates influenza haemagglutinin-specific cytotoxic T cells. Nature 311:578–579. [DOI] [PubMed] [Google Scholar]
- 71.Boliar S, Murphy MK, Tran TC, Carnathan DG, Armstrong WS, Silvestri G, Derdeyn CA. 2012. B-lymphocyte dysfunction in chronic HIV-1 infection does not prevent cross-clade neutralization breadth. J Virol 86:8031–8040. doi: 10.1128/JVI.00771-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Decker JM, Bibollet-Ruche F, Wei XP, Wang S, Levy DN, Wang W, Delaporte E, Peeters M, Derdeyn CA, Allen S, Hunter E, Saag MS, Hoxie JA, Hahn BH, Kwong PD, Robinson JE, Shaw GM. 2005. Antigenic conservation and immunogenicity of the HIV coreceptor binding site. J Exp Med 201:1407–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li B, Decker JM, Johnson RW, Bibollet-Ruche F, Wei X, Mulenga J, Allen S, Hunter E, Hahn BH, Shaw GM, Blackwell JL, Derdeyn CA. 2006. Evidence for potent autologous neutralizing antibody titers and compact envelopes in early infection with subtype C human immunodeficiency virus type 1. J Virol 80:5211–5218. doi: 10.1128/JVI.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B, Stefano-Cole K, Kuhrt DM, Gordon SN, Else JG, Mulenga J, Allen S, Sodora DL, Silvestri G, Derdeyn CA. 2010. Nonpathogenic simian immunodeficiency virus infection of sooty mangabeys is not associated with high levels of autologous neutralizing antibodies. J Virol 84:6248–6253. doi: 10.1128/JVI.00295-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lynch RM, Rong R, Boliar S, Sethi A, Li B, Mulenga J, Allen S, Robinson JE, Gnanakaran S, Derdeyn CA. 2011. The B cell response is redundant and highly focused on V1V2 during early subtype C infection in a Zambian seroconverter. J Virol 85:905–915. doi: 10.1128/JVI.02006-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Murphy MK, Yue L, Pan R, Boliar S, Sethi A, Tian J, Pfafferot K, Karita E, Allen SA, Cormier E, Goepfert PA, Borrow P, Robinson JE, Gnanakaran S, Hunter E, Kong XP, Derdeyn CA. 2013. Viral escape from neutralizing antibodies in early subtype A HIV-1 infection drives an increase in autologous neutralization breadth. PLoS Pathog 9:e1003173. doi: 10.1371/journal.ppat.1003173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rong R, Bibollet-Ruche F, Mulenga J, Allen S, Blackwell JL, Derdeyn CA. 2007. Role of V1V2 and other human immunodeficiency virus type 1 envelope domains in resistance to autologous neutralization during clade C infection. J Virol 81:1350–1359. doi: 10.1128/JVI.01839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rong R, Gnanakaran S, Decker JM, Bibollet-Ruche F, Taylor J, Sfakianos JN, Mokili JL, Muldoon M, Mulenga J, Allen S, Hahn BH, Shaw GM, Blackwell JL, Korber BT, Hunter E, Derdeyn CA. 2007. Unique mutational patterns in the envelope {alpha}2 amphipathic helix and acquisition of length in gp120 hypervariable domains are associated with resistance to autologous neutralization of subtype C human immunodeficiency virus type 1. J Virol 81:5658–5668. doi: 10.1128/JVI.00257-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rong R, Li B, Lynch RM, Haaland RE, Murphy MK, Mulenga J, Allen SA, Pinter A, Shaw GM, Hunter E, Robinson JE, Gnanakaran S, Derdeyn CA. 2009. Escape from autologous neutralizing antibodies in acute/early subtype C HIV-1 infection requires multiple pathways. PLoS Pathog 5:e1000594. doi: 10.1371/journal.ppat.1000594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Derdeyn CA, Decker JM, Bibollet-Ruche F, Mokili JL, Muldoon M, Denham SA, Heil ML, Kasolo F, Musonda R, Hahn BH, Shaw GM, Korber BT, Allen S, Hunter E. 2004. Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science 303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- 81.Ortiz AM, Klatt NR, Li B, Yi Y, Tabb B, Hao XP, Sternberg L, Lawson B, Carnathan PM, Cramer EM, Engram JC, Little DM, Ryzhova E, Gonzalez-Scarano F, Paiardini M, Ansari AA, Ratcliffe S, Else JG, Brenchley JM, Collman RG, Estes JD, Derdeyn CA, Silvestri G. 2011. Depletion of CD4(+) T cells abrogates post-peak decline of viremia in SIV-infected rhesus macaques. J Clin Invest 121:4433–4445. doi: 10.1172/JCI46023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cartwright EK, Spicer L, Smith SA, Lee D, Fast R, Paganini S, Lawson BO, Nega M, Easley K, Schmitz JE, Bosinger SE, Paiardini M, Chahroudi A, Vanderford TH, Estes JD, Lifson JD, Derdeyn CA, Silvestri G. 2016. CD8(+) lymphocytes are required for maintaining viral suppression in SIV-infected macaques treated with short-term antiretroviral therapy. Immunity 45:656–668. doi: 10.1016/j.immuni.2016.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Francella N, Gwyn SE, Yi Y, Li B, Xiao P, Elliott ST, Ortiz AM, Hoxie JA, Paiardini M, Silvestri G, Derdeyn CA, Collman RG. 2013. CD4+ T cells support production of SIV Env antibodies that enforce CD4-dependent entry and shape tropism in vivo. J Virol 87:9719–9732. doi: 10.1128/JVI.01254-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. 2006. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol 80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, Kramer VG, Siddappa NB, Xu W, Else JG, Novembre FJ, Strobert E, O'Neil SP, Ruprecht RM. 2008. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology 5:94. doi: 10.1186/1742-4690-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Shingai M, Nishimura Y, Klein F, Mouquet H, Donau OK, Plishka R, Buckler-White A, Seaman M, Piatak M Jr, Lifson JD, Dimitrov DS, Nussenzweig MC, Martin MA. 2013. Antibody-mediated immunotherapy of macaques chronically infected with SHIV suppresses viraemia. Nature 503:277–280. doi: 10.1038/nature12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Landau NR, Page KA, Littman DR. 1991. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol 65:162–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liao HX, Bonsignori M, Alam SM, McLellan JS, Tomaras GD, Moody MA, Kozink DM, Hwang KK, Chen X, Tsao CY, Liu P, Lu X, Parks RJ, Montefiori DC, Ferrari G, Pollara J, Rao M, Peachman KK, Santra S, Letvin NL, Karasavvas N, Yang ZY, Dai K, Pancera M, Gorman J, Wiehe K, Nicely NI, Rerks-Ngarm S, Nitayaphan S, Kaewkungwal J, Pitisuttithum P, Tartaglia J, Sinangil F, Kim JH, Michael NL, Kepler TB, Kwong PD, Mascola JR, Nabel GJ, Pinter A, Zolla-Pazner S, Haynes BF. 2013. Vaccine induction of antibodies against a structurally heterogeneous site of immune pressure within HIV-1 envelope protein variable regions 1 and 2. Immunity 38:176–186. doi: 10.1016/j.immuni.2012.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]