FIG 1.
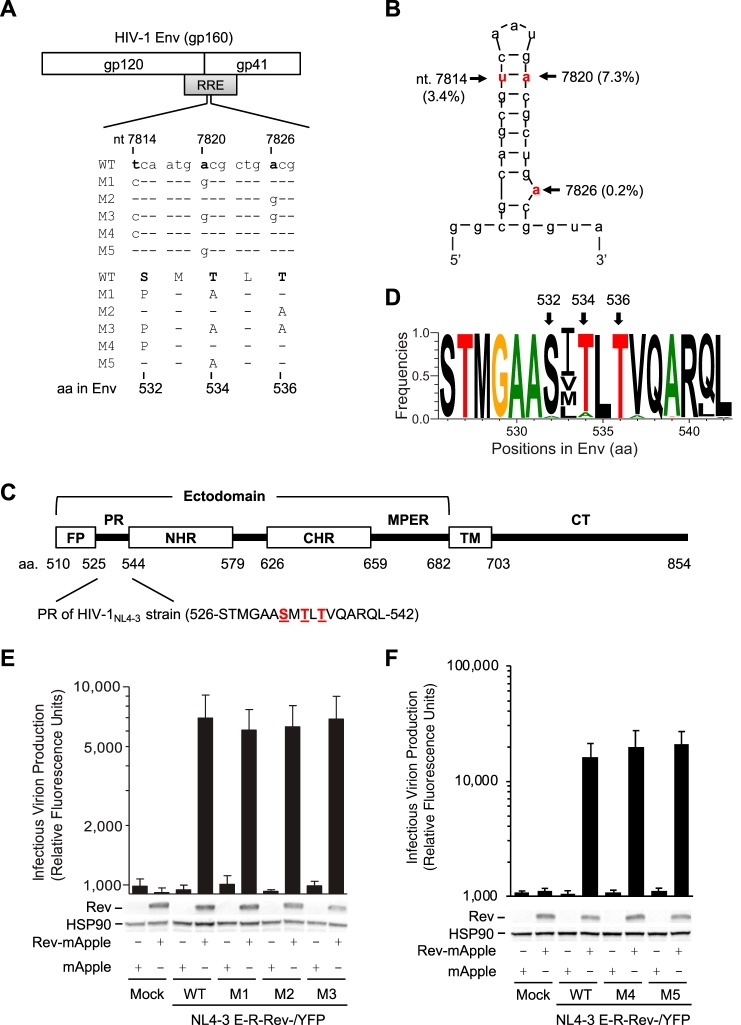
RRE mutations overlapping the gp41 coding sequence do not affect RRE function. (A) Alignment of nucleotide (nt) and amino acid (aa) sequences of overlapped HIV-1 RRE and Env in wild-type (WT) and mutants (M1 to M5) based on HIV-1NL4-3 (GenBank accession number M19921.2). (B) Secondary structure of the hairpin loop IIB of the RRE of HIV-1NL4-3. The mutation frequencies of the indicated nucleotides shown in parentheses are based on analysis of 63,612 RRE sequences in the HIV Sequence Database. Three nucleotides that were mutated are shown in red, which are equivalent to U7871, A7877, and A7883 in the HIV-1LAI strain reported by Lichinchi et al. (10). (C) HIV-1 gp41 domains (residue numbers are based on HIV-1NL4-3 strain). FP, fusion peptide; PR, polar region; NHR, N-terminal heptad repeat; CHR, C-terminal heptad repeat; MPER, membrane-proximal external region; TM, transmembrane domain; CT, cytoplasmic tail. (D) Mutation frequency of the PR amino acids in HIV-1 gp41. A total of 57,645 aligned HIV-1 Env sequences from the HIV database were analyzed. Frequency of each residue (total, 17 aa) in the PR was calculated to generate sequence logo using WebLogo. (E and F) Single-cycle HIV-1 pseudotyped with VSV-G was generated by transfecting pNL4-3 E-R-Rev-/YFP (WT or M1 to M5 separately) and plasmids encoding VSV-G or Rev fused with mApple (Rev-mApple). Plasmid encoding mApple alone was used as a negative control. Cell lysates were used for immunoblotting, and the results are shown below the bar figures (one representative from three independent experiments is shown); viruses, as indicated, were used to infect HeLa cells to quantify HIV-1 infectivity. HSP90 was used as a loading control. All experiments were performed with triplicate samples and repeated at least three times, and means ± standard errors of the means are shown. No statistical difference in levels of infectivity was observed between WT and each mutant.