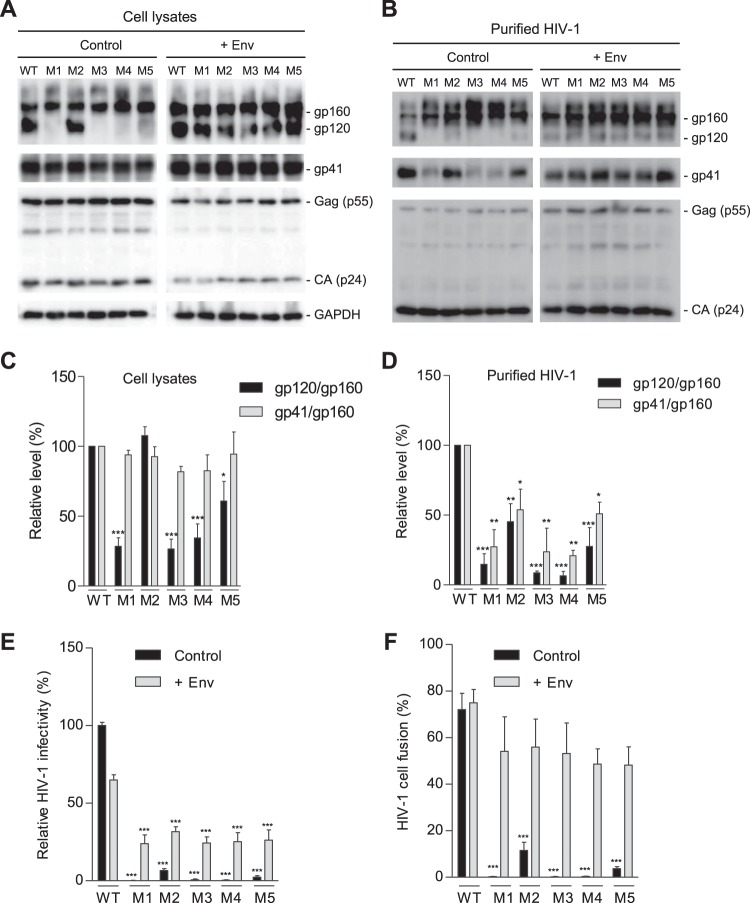
FIG 3.
PR mutations decrease gp120 association and reduce HIV-1 fusion and infectivity. HEK293T cells were transfected with full-length WT or mutant (M1 to M5) proviral constructs without (control) or with (+Env) a WT gp160 expression plasmid. (A) Cell lysates were used for immunoblotting of the indicated HIV-1 proteins. GAPDH was a loading control of cell lysates. (B) Purified HIV-1 virions were used for immunoblotting of the indicated HIV-1 proteins. (C and D) Densitometry quantification of gp160, gp120, and gp41 bands in immunoblotting of cell lysates (control samples in panel A) and purified HIV-1 (control samples in panel B), respectively. Relative levels of gp120/gp160 or gp41/gp160 ratios were calculated from three independent experiments. The values of WT HIV-1 were set as 100%, and relative levels are shown as means ± standard errors of the means (n = 3). *, P < 0.05; **, P < 0.005; ***, P < 0.0001, for the comparison of the result with an individual mutant to that with WT HIV-1 (E) WT or mutant HIV-1 generated from transfected HEK293T cells without (control) or with Env overexpression (+Env) were quantified for viral infectivity using TZM-bl cells. (F) Virion-cell fusion was determined by flow cytometry-based BlaM-Vpr assays using TZM-bl cells (10 or 50 ng of p24 for HIV-1 with or without Env trans-supplementation, respectively). All experiments were performed with triplicate samples for panels E and F and repeated at least three times, and means ± standard errors of the means are shown. Dunnett's multiple-comparison test was used for statistical analysis. ***, P < 0.0001, for the comparison of the result with an individual mutant to that with WT HIV-1.