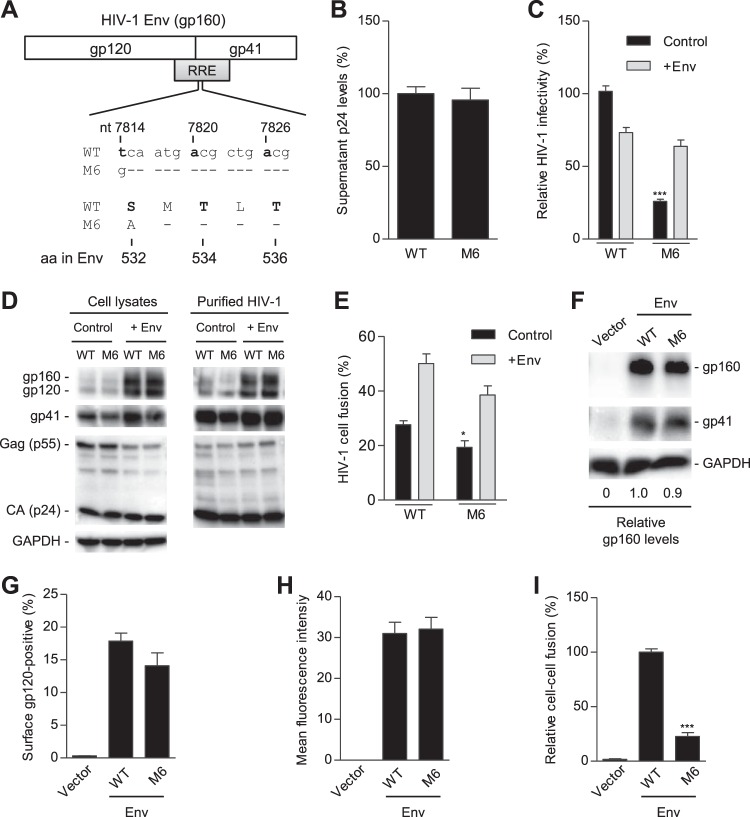
FIG 6.
S532A mutant of the PR reduces HIV-1 infectivity and fusogenicity. (A) Alignment of nucleotide (nt) and amino acid (aa) sequences of overlapped HIV-1 RRE and Env in wild-type (WT) and S532A mutants (M6) based on HIV-1NL4-3 (GenBank accession number M19921.2). (B) HIV-1 p24 levels in supernatants of transfected HEK293T cells were quantified by ELISA (n = 6). (C to E) HEK293T cells were transfected with full-length WT or M6 mutant proviral constructs without (control) or with (+Env) a WT gp160 expression plasmid. HIV-1 infectivity of equal amounts of p24 of WT and mutants was quantified with TZM-bl cells (C). (D) Lysates of transfected HEK293T cells or purified HIV-1 were analyzed for the indicated HIV-1 proteins by immunoblotting. GAPDH was a loading control (D). Virion-cell fusion was determined by flow cytometry-based BlaM-Vpr assays using TZM-bl cells (10 ng p24 for HIV-1 with or without Env trans-supplementation) (E). (F to H) HEK293T cells were separately transfected with a pRK empty vector, WT, or M6 Env (gp160) expression construct together with pCMV-Rev. HEK293T cells transfected without plasmid DNA were used as a mock control. At 24 h posttransfection, HEK293T cells were collected and lysed for immunoblotting analysis with antibodies to gp160/gp120, gp41, or GAPDH (loading control). Relative gp160 levels were quantified and normalized to the level of GAPDH (F). At 24 h posttransfection, the cells were stained with gp120 monoclonal antibody (2G12) at 4°C to measure the cell surface gp120 expression using flow cytometry. Average percentages (G) and the mean fluorescence intensity (H) of gp120-positive cells from three independent experiments were determined. (I) Transfected HEK293T cells were cocultured with TZM-bl cells for 24 h and then lysed for firefly luciferase activity measurement of Env-mediated cell-cell fusion. Average percentages of HIV-1 Env-mediated cell-cell fusion from four independent experiments are shown, with the value for the WT set as 100%. All experiments were performed with triplicate samples and repeated at least three times, and means ± standard errors of the means are shown. Dunnett's multiple-comparison test was used for statistical analysis. *, P < 0.05; ***, P < 0.0001, for results with M6 mutant compared to those with WT HIV-1.