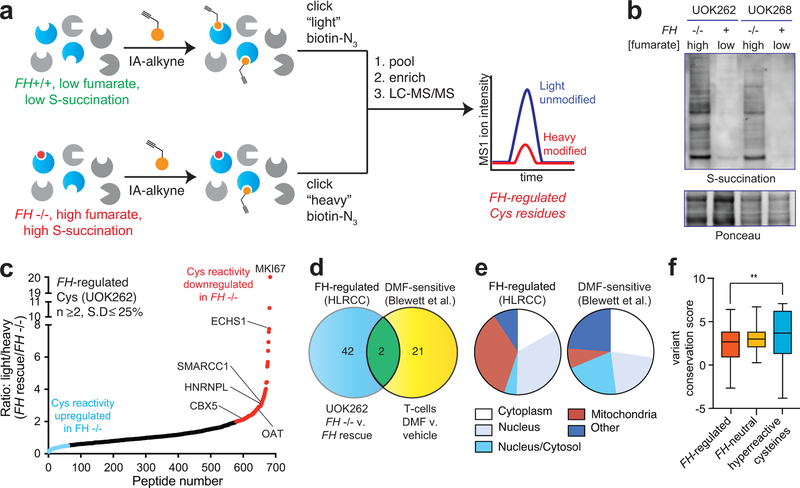
Figure 2.
Global chemoproteomic profiling of FH-regulated cysteine residues. (a) Applying a competitive chemoproteomic platform to study the oncometabolite fumarate. Comparison of proteomes from FH−/− (UOK262) and FH+/+ (UOK262WT) cells are used to define “FH-regulated” Cys residues. (b) S-succination in HLRCC cells is dependent on FH mutation. Representative image from two independent experiments is shown. Uncropped scans of immunoblot is provided in Supplementary Fig. 10. (c) FH-regulated Cys residues identified in UOK262 cells (n ≥2, SD ≤25%). (d) Overlap of FH-regulated Cys residues (R ≥2, n ≥1) with DMF-regulated Cys residues identified in Blewett et al (R ≥2, n≥1). (e) Subcellular localization of FH-regulated and DMF-sensitive Cys residues. (f) Conservation of FH-regulated, FH-neutral, and hyperreactive cysteine residues identified in Weerapana et al.16 Peptides with the highest R values from each dataset (n=50) were used for analysis. Data is presented as box and whiskers plot with box representing 25th-75th percentile, horizontal line representing median, and whiskers representing minimum and maximum values. Statistical significance was assessed using Student’s t-test (two-tailed, unpaired); **P < 0.01. Data for individual proteins is available in supplementary datasets and can be searched via a web interface at ccr2.cancer.gov/resources/Cbl/proteomics/fumarate.