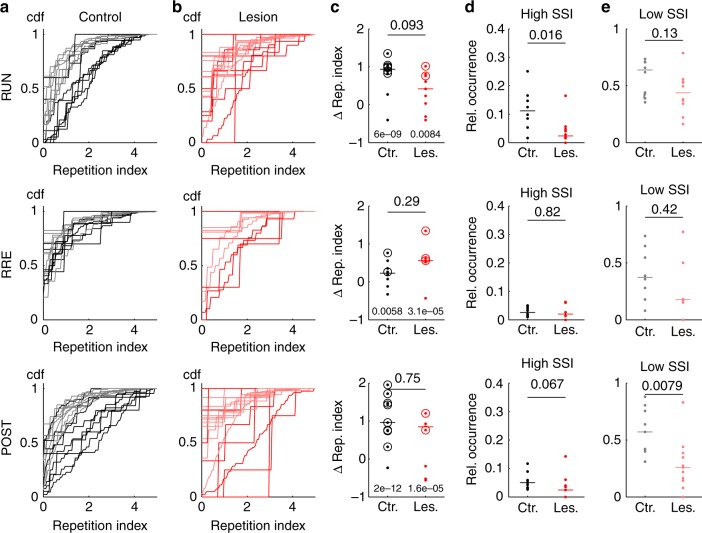
Fig. 3.
Spatial sequences recurred more frequently than nonspatial sequence motifs. a CDFs of repetition indices for spatial (significant SSI from Fig. 2b, c; black) and significant sequence motifs with low SSI (gray) in control animals for epoch types (RUN: ncontrol = 7 epochs; PRE: ncontrol = 7 epochs; POST: ncontrol = 9 epochs) as indicated. Each CDF is derived from one epoch. b Same as in (a) for MEC-lesioned animals (red: significant SSI, pink: nonsignificant SSI; RUN: nlesion = 9 epochs; PRE: nlesion = 5 epochs; POST: nlesion = 8 epochs). c Difference of medians between high- and low-SSI CDFs from (a) and (b), p values above bar were obtained from ranksum test, RUN: r.s. = 90, PRE: r.s. = 48, POST: r.s. = 85; p values above group labels were obtained from a binomial test on the number of significant epochs (circles) for a chance level of 5%; RUN: 6 out of 8 epochs were significant in control animals, 2 out of 9 epochs were significant in MEC-lesioned animals; PRE: 2 out of 8 epochs were significant in control animals, 3 out of 5 epochs were significant in MEC-lesioned animals; POST: 8 out of 9 epochs were significant in control animals, 4 out of 8 epochs were significant in MEC-lesioned animals; colors as in (a) and (b). d Relative (Rel.) occurrence computed as the fraction of spatially significant sequences among all bursts (p value obtained from ranksum test, RUN: r.s. = 103, PRE: r.s. = 62, POST: r.s. = 119; colors as in (a) and (b). e Same as d for low-SSI motifs (RUN: r.s. = 94, PRE: r.s. = 67, POST: r.s. = 130)