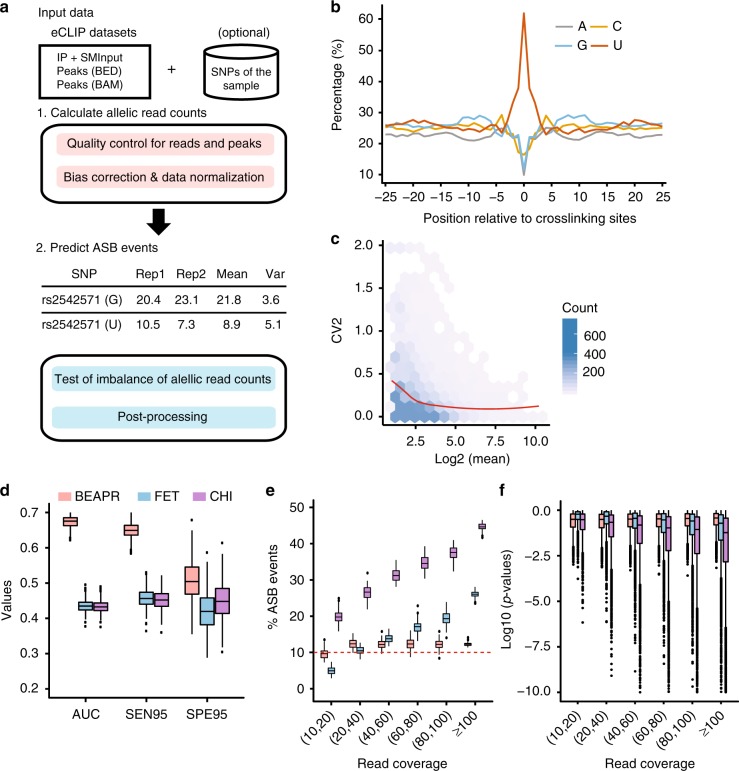
Fig. 1.
Overview of BEAPR (Binding Estimation of Allele-specific Protein–RNA interaction) and its performance. a Overall work flow of BEAPR. See Methods for details. b Crosslinking-induced bias in the SMInput sample of RBFOX2 (HepG2 cells). Y-axis shows the relative % of each nucleotide observed at each position relative to the crosslinking site (x = 0). c The square of the coefficient of variation (CV2) plotted as a function of the observed allelic read counts (mean of the two replicates) in the RBFOX2 enhanced crosslinking and immunoprecipitation (eCLIP) data in HepG2 cells (see Methods). d Performance comparison of three methods using simulated data (with simulated crosslinking-induced biases) and true allelic ratio of 0.8 for allele-specific binding (ASB). Data derived from 1000 simulation experiments each encompassing 5000 single-nucleotide variants (SNVs) (10% of which being ASB). FET: Fisher’s exact test; CHI: χ2 test; AUC: area under the curve of the precision-recall curve. SEN95: sensitivity at 95% specificity; SPE95: specificity at 95% sensitivity. e Percentage of ASB events among all tested SNVs by the three methods using simulated data as in d. The x-axis shows different read coverage bins (using average read coverage of each SNV in two simulated replicates). The red dashed line corresponds to the 10% value, that is, the percentage of true ASB events in the simulation. f Box plots of p values calculated by the three methods at different levels of read coverage. Boxplot center lines indicate the median and the boxes extend to lower and upper quartiles with whiskers depicting 1.5 interquartile range (IQR). The discrete points are the outliers. (Source data are provided as a Source Data file)