Fig. 2.
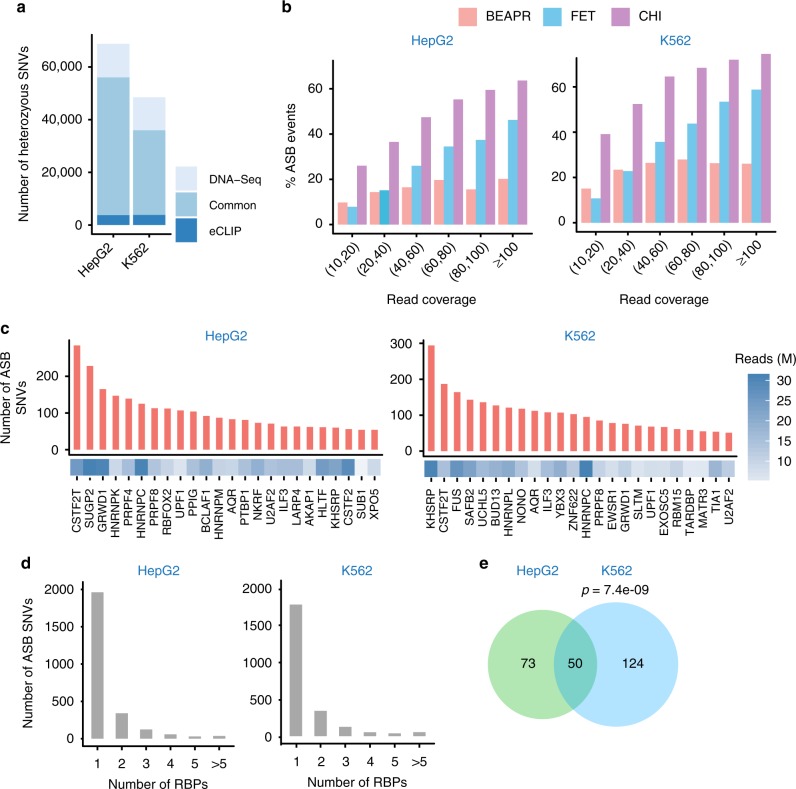
Allele-specific binding (ASB) events identified in the enhanced crosslinking and immunoprecipitation (eCLIP) data of HepG2 and K562 cells. a Number of heterozygous single-nucleotide variants (SNVs) identified via whole-genome DNA sequencing or eCLIP. Those that are common to both methods are illustrated. b Percentage of ASB events among all testable SNVs in each read coverage bin. The average read coverage of each SNV in the two replicated eCLIP experiments is shown here. A minimum read coverage of 10 was required. c Number of ASB events identified for each RNA-binding protein (RBP) in HepG2 and K562 cells. Only RBPs with ≥50 ASB events are shown. The number of usable eCLIP-Seq reads (in millions (M)) is shown for each RBP. d Number of ASB SNVs associated with one or more than one RBPs. e The overlap of ASB SNVs between HepG2 and K562. Only heterozygous SNVs common to the two cell lines are included. P value was calculated by Fisher’s exact test. (Source data are provided as a Source Data file)