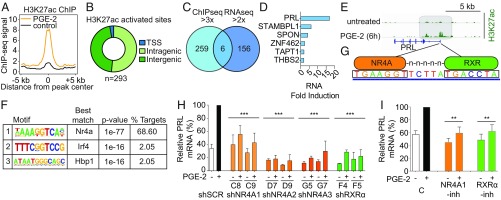
Fig. 4.
PGE2-mediated activation of prolactin in fibroblasts is mediated by NR4A-RXR heterodimers. (A) Composite plot showing genome-wide H3K27ac ChIP-Seq signals in control (black) and PGE2-treated dermal fibroblasts (DFs; orange). Data represent two biological replicates. The x axis represents a 10-kb window centered on increased H3K27ac sites in PGE2-treated cells (10 ng/mL, 6 h; n = 293 sites). (B) Distribution of increased H3K27ac sites in PGE2-treated DFs among transcriptional start sites (TSS) and intragenic and intergenic genomic loci. Promoters are annotated using the RefSeq promoter database. (C) Venn diagram comparing genes with greater than threefold increased H3K27ac activation peaks (ChIP-Seq) and those with greater than twofold up-regulated expression (RNA-Seq) in DFs after 6 h of PGE2 treatment. The 293 H3K27ac sites enriched after PGE2 treatment correspond to 265 unique genes. (D) Six genes at the intersection of the ChIP-Seq and RNA-Seq datasets are listed, with prolactin (PRL) having the highest fold induction in RNA expression in the RNA-Seq dataset. (E) ChIP-Seq data showing increased H3K27ac modification in the promoter region of the PRL gene, 6 h after PGE2 treatment of DFs. (F) Motif analysis of sites with increased H3K27ac following PGE2 treatment. The motif recognized by NR4A family members (Gene Expression Omnibus accession no. GSM777637) is the most highly enriched and accounts for the majority of sites marked by H3K27ac. (G) Nucleotide sequences between the two H3K27ac peaks in the promoter of PRL contain a potential DR5 binding site for the NR4A-RXR heterodimer. (H) Suppression of PRL mRNA induction in DFs by PGE2, following infection with shRNA constructs targeting NR4A1 (C8 and C9), NR4A2 (D7 and D9), NR4A3 (G5 and G7), RXRα (F4 and F5), or scrambled control (shSCR). Baseline and PGE2 (6 h)–induced PRL mRNA levels are shown relative to the full induction with shSCR (qPCR). Knockdown efficiency of the relevant lentiviral shRNA constructs against their targets is shown in SI Appendix, Fig. S8. Bars are the average of at least two biological replicates. Error bars are 1 SD (***P < 0.001; two-tailed Student t test; individual comparison with shSCR + PGE2). (I) Small-molecule inhibitors suppress PGE2-mediated induction of PRL mRNA. Dim-c-pPhCo2Me (10 μM) abrogates NR4A1 (39), while HX531 (1 μM) suppresses RXRα (40). Cells were treated for 18 h before PGE2 exposure. Bars represent the average of at least two biological replicates, normalized to the control PGE2-treated expression for comparison among experiments. Error bars are 1 SD (**P < 0.01, two-tailed Student t test; individual comparison with control + PGE2). C, control; -inh, inhibitor.