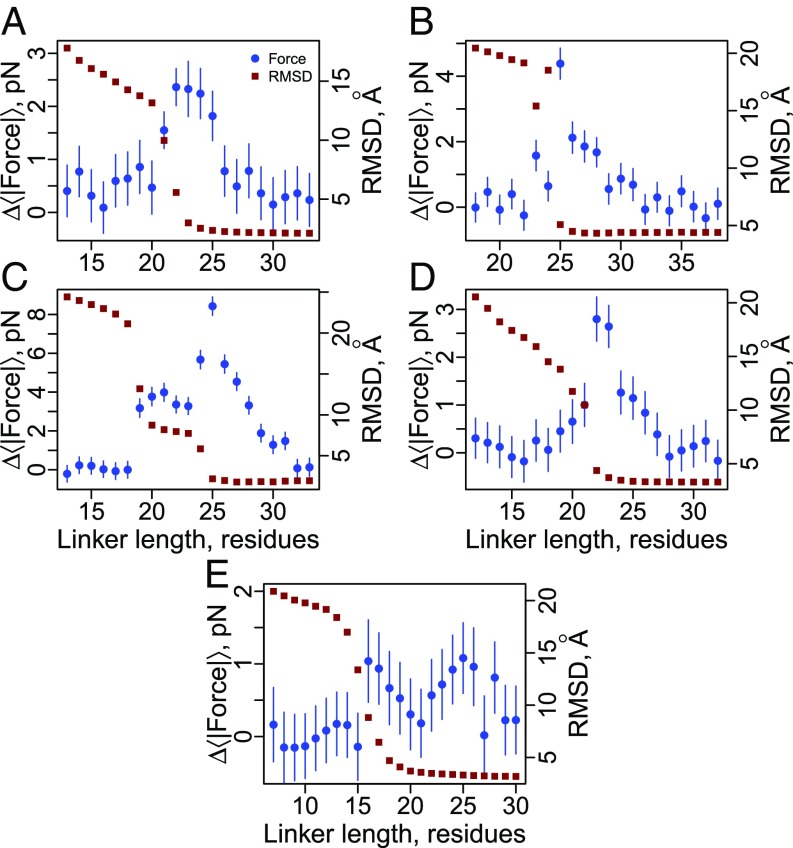
Fig. 3.
Cotranslational folding generates piconewtons of force at the P site on translationally arrested ribosomes. Blue circles correspond to the average pulling force at the C-terminal nascent chain residue (Eq. 6) as a function of linker length for the five different proteins shown in Fig. 2: (A) 1F0Z, (B) 2IST, (C) 1P9K, (D) 2JSO, and (E) 1NY8. Red squares correspond to the average rmsd difference between domain structures sampled in the simulations and the folded crystal structure. Large rmsd values indicate the domain is unfolded; low values indicate it is folded. Note that 1NY8 folds noncooperatively, and the rmsd fails to report on the docking of the C terminus beginning at a length of 25 residues, which is seen when using the fraction of native contacts per secondary structural element as an order parameter (SI Appendix, Fig. S1E). All error bars represent 95% confidence intervals about the mean calculated from block averaging. Error bars on rmsd data points are smaller than the symbols.