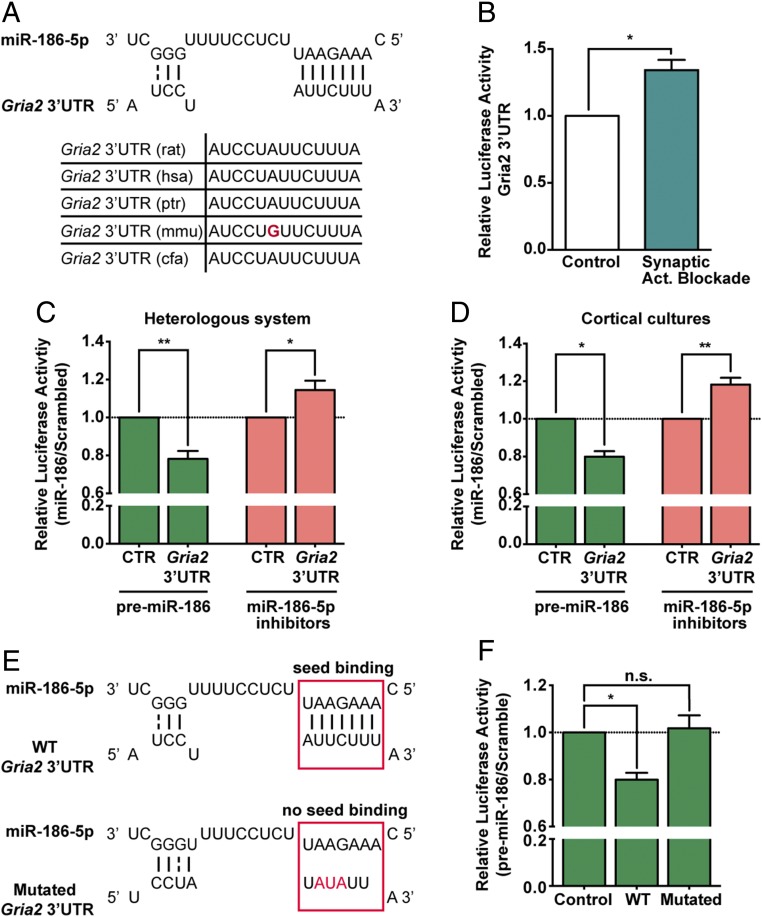
Fig. 4.
Gria2 3′UTR is a direct target of miR-186-5p. (A) miR-186-5p:Gria2 3′UTR site interaction is highly conserved in mammals. Putative site prediction performed with TargetScan and RNA hybridization analyzed with RNAhybrid. (B) Hippocampal neurons expressing guassia luciferase under the control of Gria2 3′UTR showed increased expression of guassia luciferase upon chronic blockade of synaptic activity with GYKI-52466 and MK-801 for 24 h (n = 3; one sample t test: *P ≤ 0.05). (C and D) HEK293T cells (C) or cortical neurons (D) expressing luciferase reporter constructs (pGluc-Gria2-3′UTR or pGLuc-Control) and premiR-186 or miR-186-5p inhibitors constructs (or respective scramble sequences) presented an inverse correlation between expression of miR-186-5p and luciferase under the control of Gria2 3′UTR (n = 8 or 3 for premiR-186 expression experiments and n = 6 for miR-186-5p inhibition; one sample t test: *P ≤ 0.05, **P ≤ 0.01). (E) The predicted miR-186-5p seed region targeted site in the Gria2 3′UTR was partially mutated. (F) Cortical neurons expressing luciferase constructs (control pGLuc-Control, pGLuc-GluA2-3′UTR containing the wild-type Gria2 3′UTR or pGLuc-GluA2-3′UTR containing mutated Gria2 3′UTR) and premiR-186 or scramble expressing constructs confirmed this site as a miR-186-5p target site (n = 3; one-sample t test: *P ≤ 0.05; n.s., not significant). (B–D and F) Luciferase expression was normalized for SEAP activity (expressed from the same vector) and with luciferase coexpressed with scramble controls.