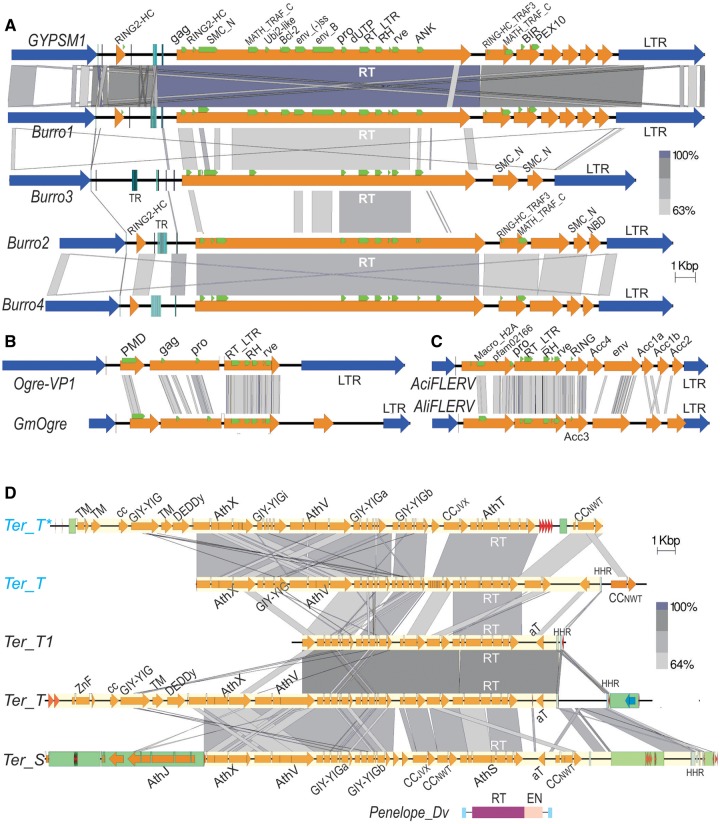
Fig. 2.
—Structure of giant retrotransposons in invertebrates. (A) Gypsy-like Burro LTR retrotransposons in the planarian Schmidtea mediterranea (Jurka 2007; Grohme et al. 2018). LTRs, blue arrows; ORFs, yellow arrows; conserved motifs mentioned in the text, green frames; vertical bars, tandem repeats (TR). (B) The longest Ogre LTR retrotransposons from soybean Glycine max (GM-Ogre) and vetch Vicia pannonica (Ogre-VP1) (Macas and Neumann 2007; Laten et al. 2008) are drawn to the same scale for comparison. (C) Foamy-like endogenous retroviruses from the midas cichlid Amphilophus citrinellus (AciFLERV) and from annual killifish Austrofundulus limnaeus (AliFLERV) (Aiewsakun and Katzourakis 2017); same scale. (D) Giant Terminon retroelements in the bdelloid rotifer Adineta vaga (Arkhipova et al. 2017); two homologous members of the same Ter_S/T family from the natural A. vaga isolate are labeled in blue. Blue vertical lines, hammerhead ribozyme motifs (HHR); green boxes, secondary TE insertions; *defective copy. For comparison, domain structure of the canonical Penelope retrotransposon from Drosophila virilis (Evgen'ev and Arkhipova 2005) is drawn to the same scale. All panels were drawn with Easyfig 2.2.2 (Sullivan et al. 2011). Regions of sequence homology are connected with gray-shaded boxes; the intensity of shading corresponds to percent BlastN (A, D) or TBlastN (B, C) identity, as indicated. Scale bar, 1 kb.