Figure 2. Analysis of the missense mutations p.Arg279Gln, p.Arg279Trp, p.Arg218Gln, and p.Val328Gly and knockdown experiments of POGLUT1 in MZ7-MEL cells.
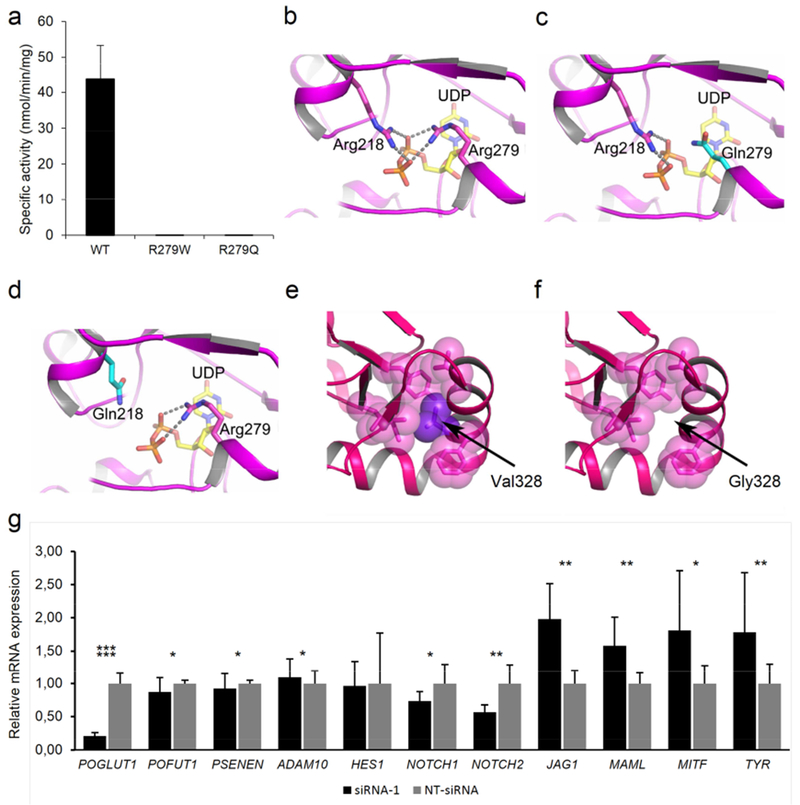
(a) Graph displaying the enzymatic activity of wild-type (WT), p.Arg279Gln, and p.Arg279Trp mutant POGLUT1 against human factor IX EGF repeat. (b-f) Protein modeling of WT and mutant POGLUT1. (b-d) The active site of POGLUT1 is shown. The residues Arg218 and Arg279 are depicted in stick representation with carbon atoms in magenta and nitrogen atoms in blue. A part of donor substrate, UDP-glucose is shown as a stick model as well, with carbon atoms in yellow, oxygen atoms in red and phosphate in orange. Both arginine residues are critical to the binding of UDP-glucose since they form salt bridges to the oxygen atoms of the UDP-glucose diphosphate moiety. The salt bridges are indicated by broken lines. Both the p.Arg218Gln and p.Arg279Gln mutation lead to substitution of a glutamine for an arginine residue (c and d, carbon atoms of Gln are depicted in cyan). Loss of the interaction between arginine and the phosphate group of UDP-glucose by substitution with glutamine will likely decrease the affinity for the substrate, and thereby the catalytic function of POGLUT1. In panel (e) the effect of mutation p.Val328Gly is modeled. The bulky sidechain of Val328 (purple) is located in a cluster of hydrophobic residues (shown in magenta as transparent spheres). (f) Mutation p.Val328Gly causes loss of this hydrophobic interaction and probably leads to the destabilization of the helix and, thus, the entire protein. (g) Analysis of siRNA-mediated POGLUT1 knockdown in MZ7-MEL cells. Relative mRNA expression levels of HES1, NOTCH1, NOTCH2, POGLUT1, POFUT1, PSENEN, ADAM10, JAG1, MAML, MITF, and TYR were evaluated by quantitative polymerase chain reaction. All experiments were carried out in triplicates. Expression levels were normalized against NT-siRNA.
