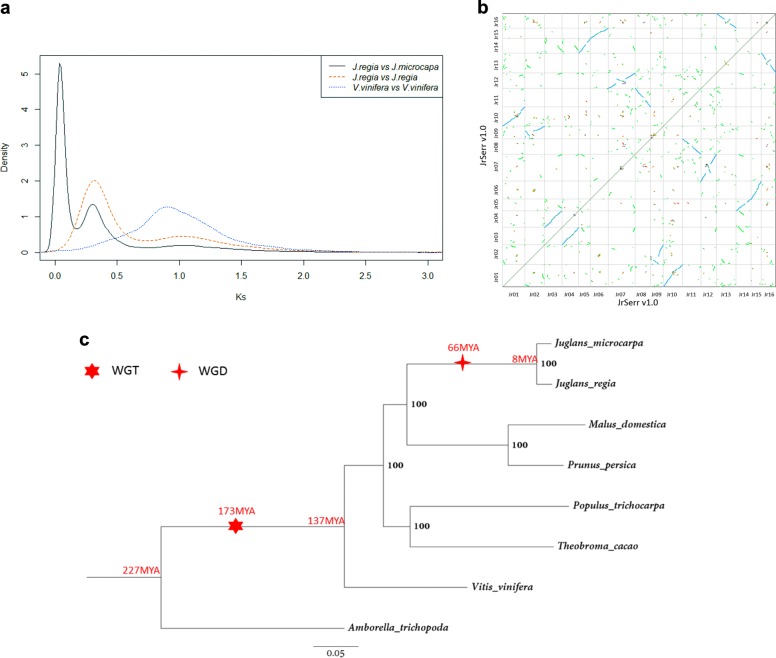
Fig. 2. Whole-genome duplication and triplication in the Juglans lineage.
a Densities of Ks divergence in the BLASTP homology searches of JrSerr_v1.0 against Jm31.01_v1.0, within JrSerr_v1.0, and within the grape genome18. b Self-alignments of the J. regia Serr pseudomolecules. Pseudomolecules are oriented as in 1b. c Divergence of the genomes of Vitis vinifera, Prunus persica, Amborella trichopoda, Populus trichocarpa, Theobroma cacao, Malus domestica, J. regia cv Serr, and J. microcarpa acc. 31.01. The dendogram was constructed from Ka divergence of 1155 single-copy genes. Bootstrap confidence on each node is shown. Also shown are estimated times of critical events in Juglans genome evolution, including the γWGT and the Juglandoid WGD (Table S13)