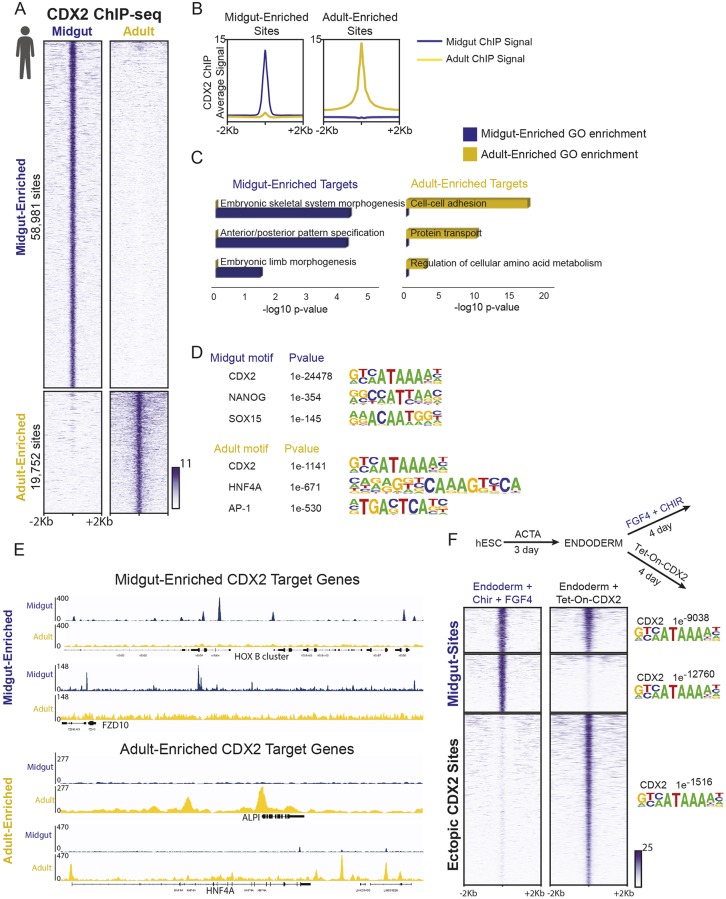
Fig. 2.
CDX2 has distinct transcriptional targets in the developing midgut versus the adult intestine. Human icon indicates data from human tissues. (A,B) CDX2 ChIP-seq performed in in vitro differentiated midgut (4 day FGF4+CHIR-treated endoderm, derived from human ESCs, as described in Fig. 1A) and in primary adult human duodenum epithelium (collected from duodenal tissue adjacent to the bile duct from patients undergoing Whipple procedures) show distinct binding profiles, as defined by k-means clustering of the identified genomic regions interacting with CDX2 (A), and compared using average signal traces at all indicated sites (B). Data from two merged biological replicates each, see Fig. S2A. (C) Distinct binding sites correspond to different sets of nearby target genes, as indicated by gene ontology (GO) analysis using DAVID (genes with their transcriptional start sites within 5 kb of CDX2-binding sites were used for analysis). (D) DNA-motif enrichment analysis finds that CDX2-binding motifs are present in both categories of binding regions, but other transcription factor-binding motifs are differentially enriched in the midgut-enriched (58,981 sites) or adult-enriched (19,752 sites) CDX2-binding regions, as defined using HOMER. (E) Examples of stage-specific CDX2-binding sites are shown using IGV to visualize merged replicates of the ChIP-seq experiments. (F) Ectopic expression of CDX2 in endoderm, using a doxycycline-inducible hESC line (Endoderm+4 days doxycycline treatment), is not sufficient to recapitulate CDX2 binding to the majority of its normal midgut target sites (Endoderm+4 days CHIR+FGF). Approximately 52% of CDX2 midgut sites (MACS P<1e−10) were not detected by MACS in the doxycycline-inducible condition (second cluster of sites in heatmap), even at a lower peak-calling stringency (P<1e−3), and 48% of the midgut regions were bound by expression of CDX2 in endoderm (first cluster of sites). Lack of binding cannot be attributed to differences in CDX2-binding motifs, which were robustly detected in each category of binding sites (HOMER). Also see Fig. S2 and Table S2.