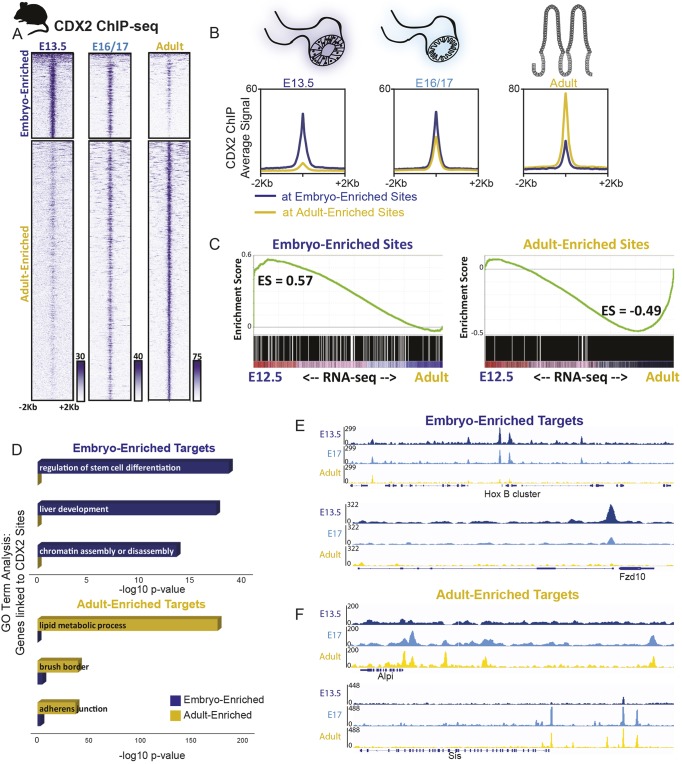
Fig. 3.
Temporal-specific shifts in CDX2 binding is conserved between mice and humans, and corresponds to temporal shifts in intestinal gene expression. Mouse icon indicates experiments performed in mouse tissues. (A) CDX2 ChIP-seq performed at the indicated stages of intestinal development on isolated epithelium show distinct, stage-dependent binding profiles (two biological replicates each, see Fig. S3; for E13.5, 12 embryos were pooled per replicate; for E16.5, three embryos pooled per replicate; for E17.5, two embryos pooled per replicate). The heatmap depicts CDX2 ChIP-seq intensity at each of the regions defined as embryo- or adult-enriched via k-means clustering of all binding sites identified in adult or embryonic conditions (Fig. S3). (B) The average ChIP-seq signals at the binding regions shown in A is plotted. (C) Embryo-enriched and adult-enriched binding sites, as identified in A, correspond to different sets of nearby target genes, which show stage-specific gene expression. RNA-seq was conducted on E12.5 or adult intestinal epithelium and genes linked to embryo-enriched or adult-enriched CDX2 binding (within 5 kb) were analyzed for their distribution along the E12.5-to-adult expression continuum using GSEA analysis. The GSEA plots indicate that embryo-enriched sites are nearby genes that are expressed at higher levels in the embryo than in the adult (leftward shift), whereas the opposite trend is observed for expression of genes nearby CDX2 sites that are stronger in the adult. CDX2-bound genes are also dependent upon CDX2 for expression, as revealed by RNA-seq analysis in Cdx2 knockout tissues (Fig. S3B). ES, enrichment score. (D) Embryo- and adult-enriched CDX2-binding regions are nearby genes with distinct gene ontologies, corresponding to embryo-specific or adult-specific functions, as indicated in GO term analysis. (E,F) Representative examples of embryo-enriched (E) or adult-enriched (F) CDX2-binding sites are shown using IGV with merged biological replicates. Also see Fig. S3.