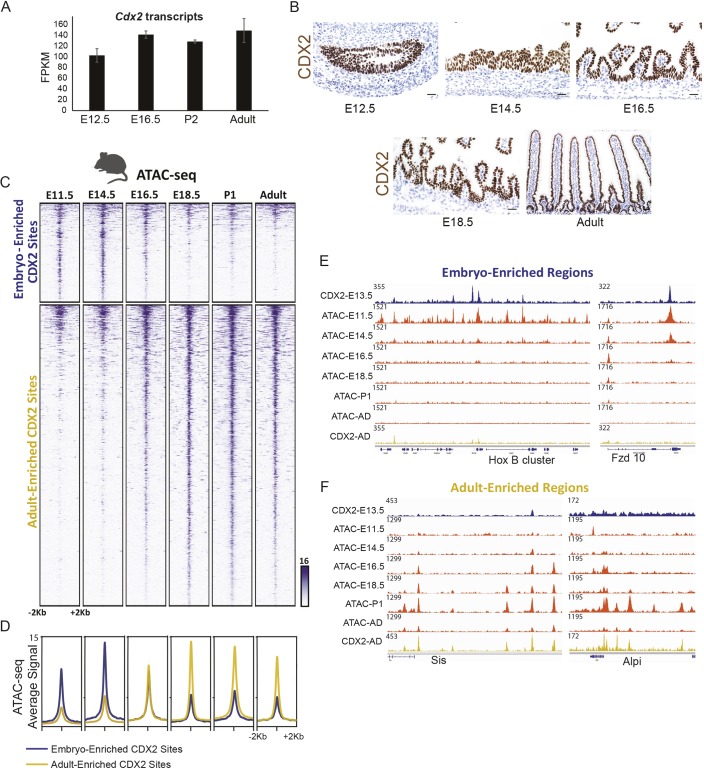
Fig. 5.
CDX2 temporally enriched binding patterns follow a temporally dynamic chromatin landscape that requires CDX2 for maintenance. (A,B) CDX2 transcripts (from RNA-seq on isolated epithelium; A) and protein levels (via immunostaining; B) are consistent across mouse developmental time, yet CDX2 occupies distinct sets of genomic regions at different developmental stages (Fig. 3). (C) CDX2 chromatin binding shifts may be due to a shifting chromatin landscape across developmental time, as indicated by the ATAC-seq assay for accessible chromatin in isolated intestinal epithelial cells. Regions shown are the CDX2 condition-enriched binding regions (as defined in Fig. 3A). (D) Average ATAC-seq signal is robust at embryo-enriched CDX2-binding sites prior to E16.5, after which chromatin accessibility at these regions diminishes. The converse is true for adult-enriched CDX2-binding sites. (E,F) Specific examples of condition-enriched chromatin accessibility at CDX2-binding sites, as visualized using IGV. CDX2-binding regions are also dependent upon CDX2 binding to remain accessible to the ATAC-seq assay, as seen in CDX2 knockout epithelium (Fig. S5).