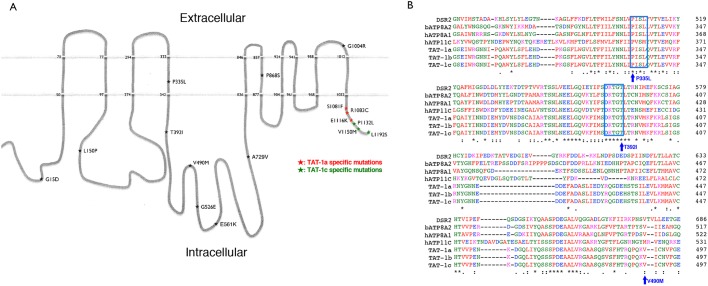
Fig. 1.
TAT-1 protein structure and location of the tat-1 mutations. (A) The schematic figure shows the structure of the TAT-1 protein with ten transmembrane domains. The positions of the analyzed TAT-1 mutations are indicated by stars. Numbers indicate mutated residues or the beginning and end of the putative transmembrane domains. The nature of the substitutions is also shown. Three mutations each alter TAT-1a and TAT-1c isoforms and are highlighted with red and green, respectively. (B) Amino acid alignment of a critical region essential for the proper TAT-1 functions. The protein sequences from three TAT-1 isoforms of C. elegans were aligned with those of P4-type ATPases from Drosophila (DSR2), cattle (ATP8A2) and humans (ATP8A1 and ATP11C). The highly conserved PISL and DKTGT motifs are shown in blue boxes. Three mutations that disrupt or reduce the functions of TAT-1 are indicated.