Abstract
Aims/hypothesis
We sought to identify the physiological implications of genetic variation at the HLA-DRB1 region in full-heritage Pima Indians in Arizona.
Methods
Single-nucleotide polymorphisms from the HLA region on chromosome 6p were tested for association with skeletal muscle mRNA expression of HLA-DRB1 and HLA- DRA, and with type 2 diabetes mellitus and prediabetic traits.
Results
The A allele at rs9268852, which tags HLA-DRB1 *02 (1602), was associated both with higher HLA-DRB1 mRNA expression (n =133, p=4.27×10−14) and decreased risk of type 2 diabetes (n=3,265, OR 0.723, p=0.002). Among persons with normal glucose tolerance (n=266) this allele was associated with a higher mean acute insulin response during an intravenous glucose tolerance test (p=0.005), higher mean 30 min insulin concentration during an oral glucose tolerance test (p=0.017) and higher body fat percentage (p=0.010). The polymorphism was not associated with HLA-DRA mRNA expression or insulin sensitivity.
Conclusions/interpretation
HLA-DRB1*02 is protective for type 2 diabetes, probably by enhancing self tolerance, thereby protecting against the autoimmune-mediated reduction of insulin secretion.
Keywords: HLA-DRB1, Insulin secretion, mRNA expression, Single-nucleotide polymorphism, Type 2 diabetes
Introduction
The Pima Indians of Arizona have a high prevalence of type 2 diabetes mellitus [1]. In 1965 a long-term study of the disease and its causes was begun by the National Institutes of Health. Research in this population has resulted in a prototypic definition of type 2 diabetes that is characterised by obesity, insulin resistance, reduced insulin secretion [2] and excess endogenous glucose production. In prospective studies, obesity, insulin resistance and the acute insulin response (AIR) predicted the disease [3, 4]. More than 2,000 Pima Indians have been characterised for HLA genotypes, which has led to the definition of new alleles at the HLA-B, HLA-C and DRB1 loci and descriptions of the distribution and population genetics of HLA variation in persons of full Pima heritage [5–7]. Recently, 100 K [8] and 1 M [9] single-nucleotide polymorphism (SNP) genome- wide association studies have been conducted in this population.
We recently completed a genome-wide gene expression study of skeletal muscle tissue samples from 133 non-diabetic Pima Indians, many of whom were part of a genome-wide 1 M SNP association study to identify genetic determinants of early onset diabetes and prediabetic traits. The expression levels of several skeletal muscle mRNA transcripts in the exon array study had a bimodal frequency distribution, and these transcripts were found to be highly significant expression quantitative trait loci (eQTL), one of which was transcribed from HLA-DRB1 [10]. The aim of the present study was to assess genetic variation at the HLA-DRB1 region and determine its possible role in gene expression and type 2 diabetes. To accomplish this we combined information from a genome-wide association study (GWAS) with serological/DNA HLA typing to identify tag SNPs and tag-haplotypes for HLA-DRB1, and we analysed the association of these variants with skeletal muscle mRNA expression, the prevalence of type 2 diabetes and prediabetic traits such as insulin secretion, thereby revealing a potential mechanism for disease protection and susceptibility.
Methods
Derivation of analytical samples
The analytical groups in Tables 1–4 and ESM Tables 1–5 were derived from five samples.
Table 1.
Genome-wide association SNP loci in the HLA region of chromosome 6 in full-heritage Pima Indians
| Locus | Position | n | Base 1, 2 | Frequency of base 1 | No. of persons by SNP genotype | ||
|---|---|---|---|---|---|---|---|
| 11 | 12 | 22 | |||||
| rs3135377 | 32385399 | 3252 | T, C | 0.003 | 0 | 19 | 3233 |
| rs9268852 | 32429594 | 3265 | A, G | 0.083 | 30 | 483 | 2752 |
| rs9268856 | 32429719 | 2907 | C, A | 0.162 | 70 | 802 | 2035 |
| rs9268858 | 32429758 | 3295 | C, T | 0.074 | 18 | 450 | 2827 |
| rs7766843 | 32538729 | 2955 | C, T | 0.151 | 63 | 769 | 2123 |
| rs9270986 | 32574060 | 3082 | A, C | 0.081 | 20 | 458 | 2604 |
| rs502771 | 32578970 | 3292 | T, C | 0.197 | 114 | 1069 | 2109 |
| rs9271720 | 32593507 | 3288 | C, T | 0.116 | 41 | 683 | 2564 |
| rs9272219 | 32602269 | 3274 | G, T | 0.069 | 17 | 415 | 2842 |
| rs9272346 | 32604372 | 3244 | G, A | 0.007 | 1 | 42 | 3201 |
| rs9272723 | 32609427 | 3222 | T, C | 0.007 | 0 | 45 | 3177 |
| rs9273363 | 32626272 | 2871 | A, C | 0.060 | 14 | 318 | 2539 |
| rs12216336 | 32967741 | 3073 | G, C | 0.123 | 57 | 644 | 2372 |
Table 4.
Reduced general linear models for the association of HLA-DRB1*02 by tag SNP rs9268852*A
| Dependent variable | n | Explanatory variable | ||||||
|---|---|---|---|---|---|---|---|---|
| DRB1 *02 | Age (years) | Sex (women) | Birth year | Body fat (%) | Insulin sensitivity | 30 min glucose | ||
| Type 2 diabetes | 3265 | 0.723a (0.002) | 1.092 (<0.001) | 1.292 (<0.001) | 1.012 (0.030) | |||
| Log acute insulin response | 266 | 1.306b (0.005) | 0.993 (0.074) | 0.807 (0.005) | 1.014 (0.006) | 0.446 (0.002) | ||
| Log 30 min insulin | 266 | 1.178c (0.017) | 0.993 (0.004) | 0.818 (0.005) | 1.021 (<0.001) | 0.299 (<0.001) | 1.007 (<0.001) | |
| Body fat percentage | 266 | 2.505d (0.010) | 0.052 (0.121) | 10.817 (<0.001) | ||||
| Log insulin sensitivity | 266 | 1.023e (0.277) | 1.000 (0.372) | 1.132 (<0.001) | 0.940 (<0.001) | |||
Each model was adjusted for the genetic correlation within sibships (p values are in parentheses)
For log-transformed variables the effect measure represents the antilog of the β coefficient
Odds ratio for diabetes in persons with presence compared with absence of the DRB1 allele
Ratio of AIR in persons with presence compared with absence of the DRB1 allele
Ratio of 30 min insulin concentration in persons with presence compared with absence of the DRB1 allele
Difference in mean body fat percentage between persons with presence or absence of the DRB1 allele
Ratio of insulin sensitivity (M) in persons with presence compared with absence of the DRB1 allele. The model included an additional covariate, body fat percentage squared, with estimate 1.000 and p=0.001
Population sample (n=3,501). A population-based sample of persons was chosen from the longitudinal study for typing with specific markers [8]. This sample consisted of all participants with available DNA whose heritage was full Pima and/or Tohono O’odham, a tribe that shares a close cultural and genetic heritage with the Pima. Examinations included measurement of venous plasma glucose concentration 2 h after the ingestion of 75 g of carbohydrate (Glucola; Ames, Elkhart, IN, USA; or Dexcola; Custom Laboratories, Baltimore, MD, USA). Diabetes was diagnosed when the 2 h post-load plasma glucose concentration was 11.1 mmol/l (200 mg/dl) or greater [11], either at a survey examination or in the course of routine medical care.
Prediabetic trait phenotype study sample (n= 266). This was a subset of the population sample with normal glucose tolerance who participated in inpatient studies to assess prediabetic traits. All of the participants were normoglycaemic by WHO 1985 criteria (i.e. fasting plasma glucose <7.8 mmol/l and 2 h post-load plasma glucose <11.1 mmol/l) [11].
Expression sample (n = 133). This was a subset of the population sample without diabetes who underwent percutaneous skeletal muscle biopsies.
GAD antibody study sample (n=208). Subjects with diabetes were selected independently of the present study to evaluate the relationship of GAD antibodies with diabetes duration and the need for insulin treatment. They were selected from four groups of participants in the longitudinal population study: newly diagnosed diabetes; long duration of diabetes not treated with insulin; long duration treated with insulin; and diabetic participants who subsequently developed end-stage renal disease from diabetic nephropathy. All individuals were full-heritage Pima.
HLA-DR sample (n=613). This was a subset of the population sample that was typed for the HLA-DR locus either by serological or high-resolution DNA methods [6, 7].
Prediabetic trait phenotype
Since 1982, non-diabetic volunteers from the Gila River Indian Community have participated in inpatient studies examining the pathophysiology of type 2 diabetes and obesity. They are admitted to the clinical research unit, where they are fed a weight-maintaining diet (50% of energy from carbohydrate, 30% from fat and 20% from protein) and abstain from strenuous exercise. After 3 days on the diet, volunteers undergo a 75 g OGTT and a series of tests including the assessment of body composition, insulin action in vivo and the AIR to intravenous glucose. Volunteers are asked to return yearly. For the present analyses, only data from the first visit with normal glucose tolerance were analysed.
Body composition was measured by underwater weighing with simultaneous determination of residual lung volume by helium dilution [12] or by total body dual energy X-ray absorptiometry (DPX-L Lunar Radiation, Madison, WI, USA) [13]. Body fat percentage was calculated; measurements using the two methods were made comparable using a previously derived equation [14]. Insulin action was assessed at physiological insulin concentrations during a hyperinsulinaemic-euglycaemic clamp [2, 15]. Briefly, after an overnight fast, a primed (1.11 MBq/min) infusion of 3-[3H]glucose infusion was started to determine the rate of postabsorptive endogenous glucose production (EGP). Two hours after starting the isotope infusion, a primed, continuous intravenous insulin infusion was administered for 100 min at a rate of 40 mU/m 2 body surface area per min. This infusion achieved a steady-state insulin concentration of (mean±SD) 876± 354 pmol/l. Plasma glucose concentrations were maintained at approximately 5.55 mmol/l with a variable infusion of 20% dextrose solution. Blood samples for measurement of 3-[3H]glucose specific activity were collected at the end of the basal period and every 10 min during the final 40 min of the insulin infusion. EGP was calculated using Steele’s equation [2, 15]. As described previously, the rate of total insulin-stimulated glucose disposal (M) was calculated for the last 40 min of the insulin infusion, which was corrected for mean glucose and insulin concentrations and EGP during the final 40 min of the insulin infusion [2, 15]. All measurements derived from the glucose clamp were normalised to estimated metabolic body size [16].
To measure the AIR, blood samples were collected before a 25 g intravenous glucose infusion over 3 min and at 3, 4, 5, 6, 8 and 10 min. The AIR was calculated as the mean increment in plasma insulin concentrations from 3 to 5 min [17].
Plasma glucose concentration was determined by the glucose oxidase method (Beckman Instruments, Fullerton, CA, USA). Plasma insulin concentrations were measured using the modification by Herbert et al. [18] of the method of Yalow and Berson [19] with an automated autoanalyser (ICN Radiochemicals, Costa Mesa, CA, USA), or using an automated immunoassay (Access, Beckman Instruments), and values from the final two assays were regressed to the original assay.
Exon array expression
Percutaneous skeletal muscle tissue biopsies were obtained from 133 non-diabetic full-heritage Pima participants over a 16-year period and stored at −70°C. Tissues were homogenised and processed with TRIzol/chloroform (Invitrogen, Carlsbad, CA, USA). Total RNA was extracted with an RNeasy Micro Kit (Qiagen, Los Angeles, CA, USA). cDNA was synthesised by reverse transcription and then hybridised with Human Exon 1.0 ST Arrays (Affymetrix, Santa Clara, CA, USA) according to the manufacturer’s protocol [Affymetrix GeneChip Whole Transcript (WT) Sense Target Labeling Assay Manual Version 4] and scanned with Affymetrix GCOS software.
Gene expression signals were normalised using the robust multichip average method [20] and GC correction. The parameters of the best-fitting bimodal distribution were estimated by maximum likelihood methods after transforming expression data to correct for skewness using the most appropriate Box-Cox parameter, as previously described [10, 21]. Individuals were classified into high and low expression categories of HLA-DRB1 based on these parameters [10].
SNP genotyping
GWAS data (1 M) were obtained as previously described [8]. On the 1 M array there were 22 SNPs with minor allele frequency >0.05 in the HLA-DRB1 region, and there were 359 individuals among those who had participated in the 1 M SNP GWAS who had also undergone serological DNA HLA typing. Seven tag SNPs were selected from among the 22 SNPs which captured the variation in all GWAS SNPs (at r2>0.8) and among the serological alleles (r2>0.6, since not all could be captured at higher r2). In addition to these tags, six SNPs that were previously reported as having strong associations with type 1 diabetes [22, 23] were selected for further genotyping. The tagger algorithm implemented in the program Haplo- view (Broad Institute, Boston, MA, USA) was used to select tags [24, 25]. The selected SNPs were genotyped in the population sample using the SNPlex genotyping System 48-plex (Applied Biosystems, Foster City, CA, USA) on an automated DNA capillary sequencer (model 3730; Applied Biosystems).
GAD antibodies
Levels of GAD antibody 65 were measured in 208 diabetic participants in this study who were also genotyped for rs9268852. GAD antibodies were measured by radioimmunoassay [26] at the University of Washington.
Informed consent
All studies were approved by the Institutional Review Board of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the Gila River Indian Community. All participants gave their informed consent.
Statistics
Haplotype frequencies for each of the serological/DNA HLA-DRB1 alleles were estimated in combination with the three SNPs that were chosen as tags for these alleles (rs9268852, rs9268858 and rs502771) These frequencies were estimated for the 613 individuals in the population with relevant data by the maximum likelihood algorithm with a filter for estimates that approximated 0.0 [27]. The four-locus haplotype frequencies were used to compute the conditional probabilities for assigning HLA- DRB1 alleles to each of the eight haplotypes that result from the three SNPs.
Tests for association were performed with a generalised linear model using the GENMOD procedure in the SAS package (SAS Institute, Cary, NC, USA). The REPEATED option was employed to account for the correlations among sibs, while the significance of the regression coefficients was calculated by comparison of each coefficient with its standard error. To estimate the odds ratio (or other relevant parameter) associated with HLA-DRB1 alleles, the DRB1 alleles were assigned at random to all persons in the sample with probability conditional on the observed tag haplotypes (or conditional on rs9268852 alone when only DRB1*02 was of interest), and this random assignment was used in the model. To account for the variation in the conditional assignment of the alleles, this was then repeated 1,000 times, and the mean of the estimates over all 1,000 replicates was taken as the parameter estimate for each model.
To account for the non-normal distribution of the GAD antibody data, differences stratified by HLA-DRB1*02 were assessed by the Wilcoxon rank-sum test.
Results
SNP rs9268852 associated with type 2 diabetes and DRB1 mRNA expression
We genotyped 13 SNPs in the HLA region of chromosome 6 in the population sample (Table 1). The hypothesis of a Hardy-Weinberg distribution of genotypes could not be rejected for any of the loci (electronic supplementary material [ESM] Table 1). Four of the SNP loci were significantly (p<0.05) associated with type 2 diabetes; the strongest association was with rs9268852, with a nominal p value of 0.004 (Table 2). The A allele at this SNP was associated with a lower prevalence of type 2 diabetes (OR 0.71). Using a dichotomised model for high and low expression, this allele was also strongly associated with higher levels of mRNA expression in skeletal muscle at the HLA-DRB1 locus (p=4.27 × 10−14; ESM Table 2).
Table 2.
Association of HLA region SNPs with type 2 diabetes, adjusted for age and sex
| SNP | Diabetes prevalence (%) by SNP genotype | ORa | 95% CI | p value | ||
|---|---|---|---|---|---|---|
| 11 | 12 | 22 | ||||
| rs3135377 | 28.7 | 43.2 | 0.53 | 0.15–1.81 | 0.3093 | |
| rs9268852 | 28.1 | 36.3 | 44.4 | 0.71 | 0.56–0.90 | 0.0038 |
| rs9268856 | 33.6 | 39.9 | 44.6 | 0.82 | 0.68–0.97 | 0.0222 |
| rs9268858 | 30.8 | 45.6 | 42.7 | 1.07 | 0.85–1.34 | 0.5694 |
| rs7766843 | 34.2 | 41.2 | 44.3 | 0.86 | 0.72–1.03 | 0.1050 |
| rs9270986 | 28.4 | 38.8 | 44.3 | 0.78 | 0.62–0.98 | 0.0345 |
| rs502771 | 36 | 41.6 | 44.1 | 0.88 | 0.76–1.03 | 0.1080 |
| rs9271720 | 27.7 | 45.4 | 42.6 | 1.02 | 0.85–1.23 | 0.8255 |
| rs9272219 | 32.6 | 44.2 | 42.9 | 1.02 | 0.80–1.28 | 0.9004 |
| rs9272346 | 100 | 22.8 | 43.3 | 0.38 | 0.16–0.93 | 0.0342 |
| rs9272723 | 25.9 | 43.5 | 0.45 | 0.20–1.02 | 0.0563 | |
| rs9273363 | 42.2 | 47.6 | 43.2 | 1.16 | 0.89–1.50 | 0.2726 |
| rs12216336 | 47.3 | 41.9 | 42.8 | 1.00 | 0.82–1.20 | 0.9620 |
Allele 1 is the reference for the odds ratio
Tag haplotypes for HLA-DRB1 alleles
Allele rs9268852*A alone acted as a tag SNP for HLA- DRB1*02 such that the probability of HLA-DRB1*02 was 0.9436 on chromosomes that carry rs9268852*A (r2=0.934; ESM Table 3). There were four common haplotypes among the three SNPs (rs9268852, rs9268858 and rs502771) that were identified as having the best potential for tags, and each of these tagged one of the common serological alleles. These four tag haplotypes defined the HLA-DRB1 variation in full- heritage Pima Indians with moderate to high probability (ESM Table 4): haplotype *A-*T-*T tagged HLA-DRB1*02 with r2=0.957; haplotype *G-*C-*T tagged HLA-DRB1*04, r 2=0.822; haplotype *G-*T-*T tagged DRB1*08, r2=0.779; and *G-*T-*C tagged DRB1*14, r2=0.630.
DRB1 tag haplotypes and DRB1 mRNA expression
In a generalised linear model DRB1 mRNA expression, as a continuous variable, was strongly and positively associated with HLA-DRB1*02 (p<0.00001; ESM Table 5). Alleles DRB1*04 and DRB1*08 and the covariates age, sex and birth year were not significantly associated with DRB1 mRNA expression in the model. There was no association of the DRB1 alleles with HLA-DRA mRNA expression in the skeletal muscle tissue samples (ESM Table 5).
DRB1 tag haplotypes, type 2 diabetes and prediabetic traits
The allele HLA-DRB1*02 was associated with a lower prevalence of type 2 diabetes (Table 3), while DRB1*04 and DRB1*08 showed no association in the model. Age-adjusted prevalence, stratified by DRB1*02, is shown in ESM Fig. 1. In 236 full-heritage Pima Indians with normal glucose tolerance, HLA-DRB1*02 was associated with a higher mean AIR during a 25 g IVGTT adjusted for, age, sex, body fat percentage and insulin sensitivity (Table 3). DRB1*02 was also associated with higher mean 30 min insulin concentration during a 75 g OGTT, adjusted for age, sex, body fat percentage, insulin sensitivity and 30 min glucose. There was no association of DRB1*04 or DRB1*08 with AIR or the mean 30 min insulin concentration (Table 3).
Table 3.
General linear models for the association of DRB1 tag haplotypes as explanatory variables with type 2 diabetes mellitus, AIR, 30 min insulin, body fat percentage and insulin sensitivity (M) as dependent variables
| Dependent variable | n | Explanatory variable | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DRB1 *02 | DRB1 *04 | DRB1 *08 | Age (years) | Sex (women) | Birth year | Body fat (%) | M | 30 min glucose | ||
| Type 2 diabetes | 2892 | 0.676a (0.001) | 0.969a (0.408) | 1.010a (0.473) | 1.087 (<0.001) | 1.225 (0.009) | 1.004 (0.282) | |||
| Log acute insulin response | 236 | 1.300b (0.010) | 0.989b (0.458) | 1.062b (0.328) | 0.986 (0.018) | 0.798 (0.006) | 1.014 (0.016) | 0.386 (0.002) | ||
| Log 30 min insulin | 236 | 1.202c (0.014) | 0.897c (0.124) | 1.099c (0.250) | 0.984 (0.001) | 0.811 (0.005) | 1.021 (<0.001) | 0.290 (<0.001) | 1.007 (<0.001) | |
| Body fat percentage | 236 | 1.967d (0.056) | −0.924d (0.194) | 0.936d (0.244) | 0.090 (0.089) | 11.166 (<0.001) | ||||
| Log insulin sensitivity | 236 | 1.054e (0.115) | 1.000e (0.490) | 1.114e (0.047) | 1.000 (0.371) | 1.138 (<0.001) | 0.948 (<0.001) | |||
Each model was adjusted for the genetic correlation within sibships (p values are in parentheses)
For log-transformed variables the effect measure represents the antilog of the β coefficient
Odds ratio for diabetes in persons with presence compared with absence of the DRB1 allele
Ratio of AIR in persons with presence compared with absence of the DRB1 allele
Ratio of 30 min insulin concentration in persons with presence compared with absence of the DRB1 allele
Difference in mean body fat percentage between persons with presence or absence of the DRB1 allele
Ratio of insulin sensitivity (M) in persons with presence compared with absence of the DRB1 allele. The model included an additional covariate, body fat percentage squared, with estimate 1.000 and p=0.008
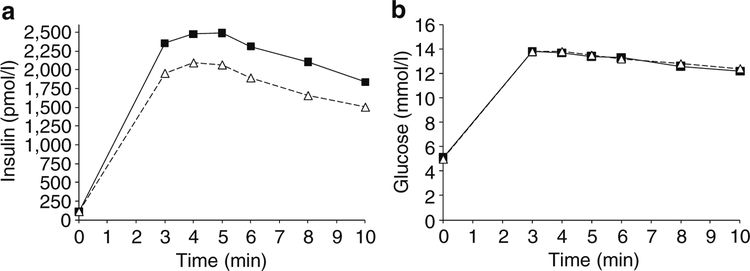
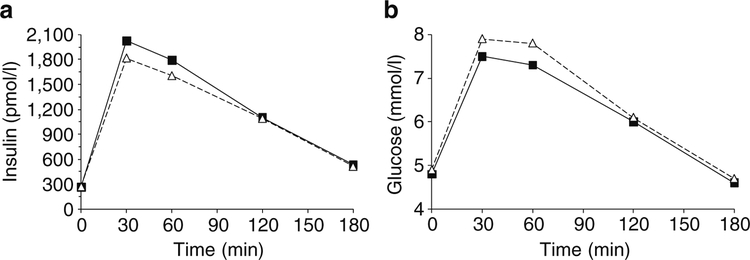
The unadjusted data for plasma insulin and glucose concentration during an IVGTT were plotted by intervals of 1 min stratified by presence/absence of the allele (Fig. 1a, b). For each of the 10 min intervals, persons with HLA- DRB1*02 had a higher mean insulin concentration, while there was no difference in mean plasma glucose concentration between persons with and without the allele. For similar unadjusted data for the OGTT test, mean plasma insulin concentrations 30 and 60 min post-load were higher for persons with DRB1*02, while their plasma glucose concen trations were lower (Fig. 2a, b). These data are consistent with higher insulin secretion in HLA-DRB1*02 carriers mediating a lower level of glycaemia after the oral load.
Fig. 1.
Mean concentrations of insulin (a) and glucose (b) in 266 participants with normal glucose tolerance stratified by the presence (squares) or absence (triangles) of HLA-DRB1*02(1602) (n=33) during an IVGTT. Insulin secretion was higher for persons with DRB1*02 during the test. After adjusting for age, sex, body fat percentage and insulin sensitivity, AIR was higher in persons with DRB1*02 (p=0.005, Table 4)
Fig. 2.
Plasma concentrations of insulin (a) and glucose (b) after a 75 g OGTT in 266 participants with normal glucose tolerance stratified by the presence (squares) or absence (triangles) of HLA-DRB1* 02(1602). After adjusting for covariates age, sex, body fat percentage, 30 min glucose and insulin sensitivity, the 30 min insulin concentration was higher (p=0.017, Table 4) for those with DRB1*02
There was no association between insulin sensitivity and imputed DRB1 alleles, as assessed by a hyperinsulinaemic- euglycaemic clamp (M). However, there was a nominally significant (p=0.047) association with HLA-DRB1*08, such that carriers tended to be more insulin sensitive.
Imputation of HLA-DRB1*02 by tag SNP rs9268852 alone
With the exception of DRB1*08 and insulin sensitivity, alleles DRB1*04 and DRB1*08 were not significantly related to the dependent variables in Table 3; the analyses were therefore repeated with DRB1*02 assigned by the tag SNP rs9268852*A alone. Since only a single SNP was required for these analyses, the number of persons available for assignment in the model increased, for type 2 diabetes, from 2,892 to 3,265 and, for prediabetic traits, from 236 to 266. Similar statistically significant results were obtained (Table 4). In addition, there was an association between HLA-DRB1*02 and body fat percentage adjusted for age and sex, such that carriers of HLA-DRB1*02 had higher levels of body fat (despite the lower risk of diabetes) (Table 4).
Imputed DRB1 alleles and GAD antibodies
Patients with diabetes who were DRB1*02+ had a lower mean level of GAD antibodies (n=38, mean antibody units 0.038 vs DRB1*02-, n =170, 0.061 antibody units). The antibody level was ≥0.15 antibody units in 10 (6.0%) of the 170 participants without DRB1*02 (ESM Fig. 2), but in none of the 38 participants carrying the allele. However, the Wilcoxon two-sample test for the difference in distributions was not significant (p=0.143).
Discussion
We found that the A allele at rs9268852 was associated with higher expression of DRB1 mRNA, lower prevalence of type 2 diabetes and increased insulin secretion in response to an intravenous and oral glucose load. In addition, while not reaching statistical significance at p < 0.05, the mean level of GAD antibodies was lower, which is consistent with a previous report in a large number of Japanese people with age at onset >20 years [28]. In the Pima population rs9268852*A is highly concordant with HLA-DRB1*02. From a previous study of the high- resolution HLA variation in full-heritage Pimans, it is known that the molecular allele is HLA-DRB1*1602 [7].
We hypothesise from these data that the HLA-DRB1*02 (1602) allele reduces the risk of type 2 diabetes because it confers greater ability to maintain self tolerance during ageing and therefore protects from an autoimmune-mediated reduction in insulin secretory function, either by a loss of beta cell mass or interference with early insulin release. This would also explain the lower level of GAD antibodies. The association of HLA-DRB1* 1602 with increased mRNA expression of locus DRB1 would also be consistent with this hypothesis if there were a protective dosage effect in antigen-presenting cells. Increased expression of the HLA- DR heterodimer with the DRB1*1602 β chain might lead to a higher density of peptide-presenting molecules on the surface of the cell that are available to T cell recognition and thereby amplify the maintenance of self tolerance.
de Bakker et al. [29] reported that the best tag SNP for HLA-DRB1*02(1502) in persons of European descent is rs3135388, which is highly concordant in HapMap Phase II with rs9270986. Locus rs9270986 is strongly associated with type 1 diabetes [22] and with type 2 diabetes in the Diabetes Genetics Replication and Meta-analysis Consortium (DIAGRAM) study [30, 31]. In Pimas we found that rs9270986 was also associated with diabetes (Table 2) and was strongly concordant with rs9268852, our primary DRB1*02 tag. Haplotype rs9270986*A-DRB1*02 exhibited strong, positive genetic disequilibrium in Pima Indians (n= 732, D=0.075, D’=0.983). A meta-analysis for rs9270986 (R. Hanson, unpublished results), which combines the Pimas with DIAGRAM, gives a pooled (n =13,210) OR of 0.83 (95% CI 0.77–0.90) and improves the evidence for association by an order of magnitude (p=5.5×10−6). Although this does not achieve conventional thresholds for genome-wide significance, this is nonetheless strong evidence of association, particularly given that this SNP has one of the strongest associations with type 1 diabetes [22]. While the association of type 2 diabetes with HLA variants in the Europeans in DIAGRAM might be attributable to misclassification of type 1 diabetes, the similar association in Pimas makes this less likely as type 1 diabetes is virtually absent in this population [32, 33].
Since 1977 [34], the inverse association between HLA- DRB1*02 and type 1 diabetes has been reported in many different ethnic groups [35–38]. The investigators of the Type 1 Diabetes Genetics Consortium recently reported that persons with HLA-DRB1*02 are protected from the disease [39]. The high-resolution molecular subtypes of DRB1*02, *1501, *1502 (in European populations) and now *1602 all share the property of protection, not risk. It is possible, therefore, that protection from diabetes is a property common to all of the HLA-DR2 functional DRA1-DRB1 heterodimer molecules and not the unique property of one high-resolution allelic subtype of HLA-DRB1*02.
In persons with type 1 disease a well-defined spectrum of autoantigens, including GAD, contribute to the loss of self tolerance and autoimmune diabetes mellitus [40, 41], the highest risk factor being the number of kinds of autoantibodies [42]. In Finland, GAD antibodies were predictive of insulin deficiency [43]. It has also been shown that DRB1*02, either *1501 or *1502, can moderate the effect of extreme levels of autoantibodies to GAD [44].
There is no direct experimental evidence of loss of self tolerance with ageing that is arrested by allele DRB1*02. However, a mechanism for this protection is suggested by transgenic mouse experiments in which the human HLA- DRB1 and/or DQB1 alleles are spliced into the mouse genome, after which the animals are injected with rat GAD antigen, an autoantigen homologous to that of the mouse. The presence of protective HLA-DRB1*02(1502) prevented the generation of a self-reactive response and insulitis, whereas splicing of the susceptible allele DRB1*0301 generated a response to the homologous rat GAD protein [45, 46].
Schadt et al. [47] reported a strong association between SNP rs9272723 and expression of the nearby gene HLA- DRB1 in a study of 400 human liver samples. Zhong et al. [48] recently combined data from genome-wide association and mRNA expression studies and identified HLA-DRB1 as an important antigen processing and presentation pathway in type 2 diabetes. We know of no previous data on the relationship between an HLA-DRB1 allele and mRNA expression.
The major strength of the present work is the combination of clinical and physiological data with measures of gene expression and HLA SNP genotyping in a culturally and genetically homogeneous population. A weakness of the study is the relatively small number of participants, which leads us to treat the significance levels of the analyses with caution and to emphasise the need for replication in other populations.
The decreased risk of diabetes in people who have DRB1*02 in this population may raise the question whether the diabetes that occurs is a form of latent autoimmune diabetes of adults (LADA). Generally accepted diagnostic criteria for LADA, as developed by the Immunology of Diabetes Society, are onset above the age of 30 years, the presence of autoantibodies and no requirement for insulin treatment for at least 6 months after diagnosis [49]. Patients with LADA are also often lean [49]. The clinical phenotype of diabetes in the Gila River Indian Community is more consistent with that of type 2 diabetes, i.e. affected individuals are obese, insulin resistant, as well as having reduced insulin secretion, and often go for years without an absolute requirement for insulin therapy [1]. The diagnosis of LADA is not appropriate for most people with diabetes in this population and we doubt the utility of this definition, as suggested by others [50]. The important implication from the present work is that there appears to be an autoimmune- mediated loss of insulin secretion in those who do not have DRB1*02, apparent even in young individuals with normal glucose tolerance. This loss of insulin secretory function increases the risk of diabetes—the categorical definition of which is most consistent with type 2 diabetes mellitus.
Identifying the mechanism of HLA-DRB1*02 protection from type 2 diabetes is an important area for future research. If a gradual loss of self tolerance during ageing contributes to susceptibility to type 2 diabetes, then methods need to be developed to monitor the incidence of autoimmune antibodies, their nature and target, and their strength. This study provides compelling evidence that the two major categories of diabetes, type 1 and type 2, are in many cases a manifestation of a more general syndrome, which has a common association with HLA class II loci. The protective associations of HLA-DRB1*02 with type 1 and type 2 diabetes and, in particular, insulin secretion, offer a direction for future research to define the detailed mechanics of HLA in protection from and susceptibility to diabetes in humans.
Acknowledgements
We thank the research volunteers for their assistance and the staff of the Phoenix Epidemiology and Clinical Research Branch, NIDDK, for conducting the examinations. This research was partially supported by grants BSF45–3 and BSF45–4 from Blood Systems Foundation, Blood Systems, Scottsdale, AZ, USA, by the Intramural Research Program of the NIDDK, and grants 7–04-DCS-02 and 7–06-MN-06 to C. Bogardus from the American Diabetes Association.
Abbreviations
- AIR
Acute insulin response
- EGP
Endogenous glucose production
- GAD
Glutamic acid decarboxylase
- GWAS
Genome-wide association study
- SNP
Single-nucleotide polymorphism
Footnotes
Electronic supplementary material The online version of this article (doi:10.1007/s00125-011-2122-8) contains supplementary material, which is available to authorised users.
Duality of interest The authors declare that there is no duality of interest associated with this manuscript.
Contributor Information
R. C. Williams, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
Y. L. Muller, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
R. L. Hanson, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
W. C. Knowler, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
C. C. Mason, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
L. Bian, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
V. Ossowski, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
K. Wiedrich, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
Y. F. Chen, Human Development, Tzu Chi University, Hualien, Taiwan
S. Marcovina, Northwest Lipid Metabolism and Diabetes Research Laboratories, Seattle, WA, USA
J. Hahnke, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
R. G. Nelson, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
L. J. Baier, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
C. Bogardus, Phoenix Epidemiology and Clinical Research Branch, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, 1550 East Indian School Road, Phoenix, AZ 85014, USA, williamsr@mail.nih.gov
References
- 1.Knowler WC, Pettitt DJ, Saad MF, Bennett PH (1990) Diabetes mellitus in the Pima Indians: incidence risk factors and pathogenesis. Diab Metab Rev 6:1–27 [DOI] [PubMed] [Google Scholar]
- 2.Lillioja S, Mott DM, Spraul M et al. (1993) Insulin resistance and insulin secretory dysfunction as precursors of non-insulin dependent diabetes mellitus: prospective studies in Pima Indians. N Engl J Med 329:1988–1992 [DOI] [PubMed] [Google Scholar]
- 3.Weyer C, Tataranni PA, Bogardus C, Pratley RE (2001) Insulin resistance and insulin secretory dysfunction are independent predictors of worsening of glucose tolerance during each stage of type 2 diabetes development. Diab Care 24:89–94 [DOI] [PubMed] [Google Scholar]
- 4.Weyer C, Bogardus C, Mott DM, Pratley RE (1999) The natural history of insulin secretory dysfunction and insulin resistance in the pathogenesis of type 2 diabetes. J Clin Invest 104:787–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams RC, McAuley JE (1992) HLA class I variation controlled for genetic admixture in the Gila River Indian Community of Arizona: a model for the Paleo-Indians. Hum Immunol 33:39–46 [DOI] [PubMed] [Google Scholar]
- 6.Williams RC, McAuley JE (1992) HLA class II variation in the Gila River Indian Community of Arizona: alleles, haplotypes, and a high frequency epitope at the HLA-DR locus. Hum Immunol 33:29–38 [DOI] [PubMed] [Google Scholar]
- 7.Williams RC, Chen YF, Endres RO et al. (2009) Molecular variation at the HLA-A, B, C, DRB1, DQA1, and DQB1 loci in full heritage American Indians in Arizona: private haplotypes and their evolution. Tissue Antigens 74:520–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson RL, Bogardus C, Duggan D et al. (2007) A search for variants associated with young-onset type 2 diabetes in American Indians in a 100K genotyping array. Diabetes 56:3045–3052 [DOI] [PubMed] [Google Scholar]
- 9.Hanson RL, Knowler WC, Bogardus C, Bian L, Kobes S, Baier LJ (2010) Variants associated with young-onset type 2 diabetes in American Indians among 454,194 single nucleotide polymorphisms (SNPs). Diabetes 59(Suppl 1A):LB18 [Google Scholar]
- 10.Mason CC, Hanson RL, Ossowski V et al. (2011) Bimodal distribution of RNA expression levels in human skeletal muscle tissue. BMC Genomics 12:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Diabetes Mellitus: Report of a WHO Study Group (1985) Technical Report Series 727. World Health Organization, Geneva: [PubMed] [Google Scholar]
- 12.Goldman RF, Buskirk ER (1961) A method for underwater weighing and the determination of body density In: Brozek J, Herschel A (eds) Techniques for measuring body composition. National Academy of Sciences, Washington, pp 78–106 [Google Scholar]
- 13.Mazess RB, Barden HS, Bisek JP, Hanson J (1990) Dual-energy X-ray absorptiometry for total-body and regional bone- mineral content and soft-tissue composition. Am J Clin Nutr 51:1106–1112 [DOI] [PubMed] [Google Scholar]
- 14.Tataranni PA, Ravussin E (1995) Use of dual-energy X-ray absorptiometry in obese individuals. Am J Clin Nutr 62:730–734 [DOI] [PubMed] [Google Scholar]
- 15.Bogardus C, Lillioja S, Mott D, Reavon JR, Kashiwagi A, Foley JE (1984) Relationship between obesity and maximal stimulated glucose uptake in vivo and in vitro in Pima Indians. J Clin Invest 78:1568–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lillioja S, Bogardus C (1988) Obesity and insulin resistance: lessons learned from the Pima Indians. Diab Metab Rev 4:517–540 [DOI] [PubMed] [Google Scholar]
- 17.Schwartz MW, Boyko EJ, Kahn SE, Ravussin E, Bogardus C (1995) Reduced insulin secretion: an independent predictor of body weight gain. J Clin Endocrinol Metab 80:1571–1576 [DOI] [PubMed] [Google Scholar]
- 18.Herbert Y, Lau K, Gottlieb CW, Bleicher SJ (1965) Coated charcoal immunoassay of insulin. J Clin Endocrinol Metab 25:1375–1385 [DOI] [PubMed] [Google Scholar]
- 19.Yalow RS, Berson SA (1960) Immunoassay of endogenous plasma insulin in man. J Clin Invest 39:1157–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Irizarry RA, Hobbs B, Collin F et al. (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4:249–264 [DOI] [PubMed] [Google Scholar]
- 21.MacLean CJ, Morton NE, Elston RC, Yee S (1976) Skewness in commingled distributions. Biometrics 32:695–699 [PubMed] [Google Scholar]
- 22.Nejentsev S, Howson JM, Walker NM et al. (2007) Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Todd JA, Walker NM, Cooper JD et al. (2007) Robust associations of four new chromosome regions from genome-wide analyses of type 1 diabetes. Nat Genet 39:857–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bakker PIW, Yelensky R, Pe’er I, Gabriel SB, Daly MJ, Altshuler D (2005) Efficiency and power in genetic association studies. Nat Genet 37:1217–1223 [DOI] [PubMed] [Google Scholar]
- 25.Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization ofLD and haplotype maps. Bioinformatics 21:263–265 [DOI] [PubMed] [Google Scholar]
- 26.Falorni A, Ortqvist E, Persson B, Lernmark A (1995) Radio-immunoassays for glutamic acid decarboxylase (GAD65) and GAD65 autoantibodies using 35S or 3H recombinant human ligands. J Immunol Meth 186:89–99 [DOI] [PubMed] [Google Scholar]
- 27.Long JC, Williams RC, Urbanek M (1995) An EM algorithm and testing strategy for multiple locus haplotypes. Am J Hum Genet 56:799–810 [PMC free article] [PubMed] [Google Scholar]
- 28.Takeda H, Kawasaki E, Shimizu I et al. (2002) Clinical, autoimmune, and genetic characteristics of adult-onset diabetic patients with GAD autoantibodies in Japan (Ehime Study). Diab Care 25:995–1001 [DOI] [PubMed] [Google Scholar]
- 29.de Bakker PIW, McVean G, Sabeti PC et al. (2006) A high resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38:1166–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeggini E, Scott LJ, Saxena R et al. (2008) Meta-analysis of genome-wide association data and large scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 40:638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voight BF, Scott LJ, Steinthorsdottir V et al. (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association studies. Nat Genet 42:579–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Savage PJ, Bennett PH, Senter RG, Miller M (1979) High prevalence of diabetes in young Pima Indians. Diabetes 28:937–942 [DOI] [PubMed] [Google Scholar]
- 33.Dabalea D, Palmer JP, Bennett PH, Pettitt DJ, Knowler WC (1999) Absence of glutamic acid decarboxylase antibodies in Pima Indian children with diabetes mellitus. Diabetologia 42:1265–1266 [DOI] [PubMed] [Google Scholar]
- 34.Batchelor JR, Morris PJ (1977) HLA and disease In: Bodmer WF, Batchelor JR, Bodmer JG, Festenstein H, Morris PJ (eds) Histocompatibility testing 1977. Munksgaard, Copenhagen, pp 205–258 [Google Scholar]
- 35.Tiwari JL, Terasaki PI (1985) HLA and disease associations. Springer, New York [Google Scholar]
- 36.Zhang H, Wang B, Zhao X et al. (2000) The susceptible alleles on HLA-DRB1 of type 1 diabetes in children in Harbin. Zhonghua Liu Xing Bing Xue Za Zhi 21:267–269 [PubMed] [Google Scholar]
- 37.Song D, Liu Y, Han Y et al. (2002) Study on the gestational diabetes mellitus and histocompatibility human leukocyte antigen DRB allele polymorphism. Zhonghua Fu Chan Ke Za Zhi 37:284–286 [PubMed] [Google Scholar]
- 38.Zhang XM, Wang HY, Luo YY, Ji LN (2009) HLA-DQ, DR allele polymorphism of type 1 diabetes in the Chinese population: a meta-analysis. Chin Med J Engl 122:980–986 [PubMed] [Google Scholar]
- 39.McKinnon E, Morahan G, Nolan D, James I, Type 1 Diabetes Genetics Consortium (2009) Association of MHC SNP genotype with susceptibility to type 1 diabetes: a modified survival approach. Diabetes Obes Metab 11:92–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Taplin C, Barker JM (2008) Autoantibodies in type 1 diabetes. Autoimmunity 41:11–18 [DOI] [PubMed] [Google Scholar]
- 41.Knip M, Siljander H (2008) Autoimmune mechanisms in type 1 diabetes. Autoimmun Rev 7:550–557 [DOI] [PubMed] [Google Scholar]
- 42.Betterle C, Spadaccino C, Presotto F et al. (2002) The number of markers of pancreatic autoimmunity is proportional to the risk for type 1 diabetes mellitus in Italian and English patients with organ- specific autoimmune diseases. Ann NY Acad Sci 958:276–280 [DOI] [PubMed] [Google Scholar]
- 43.Niskanen LK, Tuomi T, Karjalainen J, Groop LC, Uusitupa MIJ (1995) GAD antibodies in NIDDM. Diab Care 18:1557–1565 [DOI] [PubMed] [Google Scholar]
- 44.Ishii M, Hasegawa G, Fukui M et al. (2005) Clinical and genetic characteristics of diabetic patients with high-titer (>10,000 U/ml) of antibodies to glutamic acid decarboxylase. Immunol Lett 99:180–185 [DOI] [PubMed] [Google Scholar]
- 45.Abraham RS, Kudva YC, Wilson SB, Strominger JL, David CS (2000) Co-expression of HLA-DR3 and DQ8 results in the development of spontaneous insulitis and loss of tolerance to GAD65 in transgenic mice. Diabetes 49:548–554 [DOI] [PubMed] [Google Scholar]
- 46.Abraham RS, Wen L, Marietta EV, David CS (2001) Type 1 diabetes-predisposing MHC alleles influence the selection of glutamic acid decarboxylase (GAD) 65-specific T cells in a transgenic model. J Immunol 166:1370–1379 [DOI] [PubMed] [Google Scholar]
- 47.Schadt EE, Molony C, Chudin E et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6:1020–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong H, Yang X, Kaplan LM, Molony C, Schadt EE (2010) Integrating pathway analysis and genetics of gene expression for genome-wide association studies. Am J Hum Genet 86:581–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Naik RG, Brooks-Worrell BM, Palmer JP (2009) Latent autoimmune diabetes in adults. J Clin Endocrinol Metab 94:4635–4644 [DOI] [PubMed] [Google Scholar]
- 50.Gale EAM (2005) Latent autoimmune diabetes in adults: a guide for the perplexed. Diabetologia 48:2195–2199 [DOI] [PubMed] [Google Scholar]