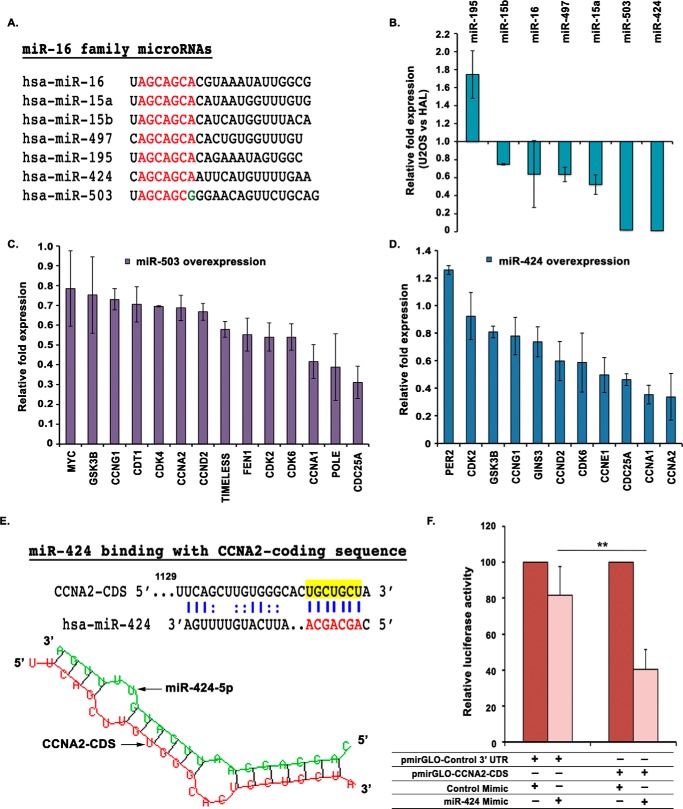
Figure 6.
miR-424, a miR-16 family miRNA, targets CCNA2 and is down-regulated in aggressive osteosarcoma. A, alignment of the mature sequences of miR-16 family miRNAs; the seed sequences of all miRNAs are highlighted in red. miR-503 is shown along with miR-16 family, as their seed sequences are very similar. B, relative expression of all mature miRNAs of miR-16 family in rapid-proliferating osteosarcoma cell line, U2OS, compared with slow-proliferating cell line, HAL. The expression was determined by qRT-PCR and normalized with the expression of small nucleolar RNA, RNU48. C and D, relative expression of putative target genes after three consecutive transfections of U2OS cells with either miR-503 or miR-424 mimic. The -fold expression of target genes is represented compared with control mimic–transfected cells and normalized with the expression of the β2-microglobulin gene. For expression levels of miRNA/mRNA, the data are represented as the mean of two experiments, each with two technical replicates, ± S.D. (error bars). E, an illustration of miR-424 binding within the CCNA2-coding sequence, which was cloned downstream of the firefly luciferase gene in pmirGLO Dual-Luciferase vector. miR-424 binding to CCNA2 as per RNAhybrid has been depicted. F, U2OS cells were transfected with pmirGLO Dual-Luciferase vector in which either the coding sequence of CCNA2 or a nonspecific 3′-UTR has been cloned downstream of the firefly luciferase gene. The cells were co-transfected with either miR-424 or control mimic, and the relative firefly/Renilla luciferase activity was determined for each sample. Data are represented as the mean of four experiments ± S.D. **, p < 0.01.