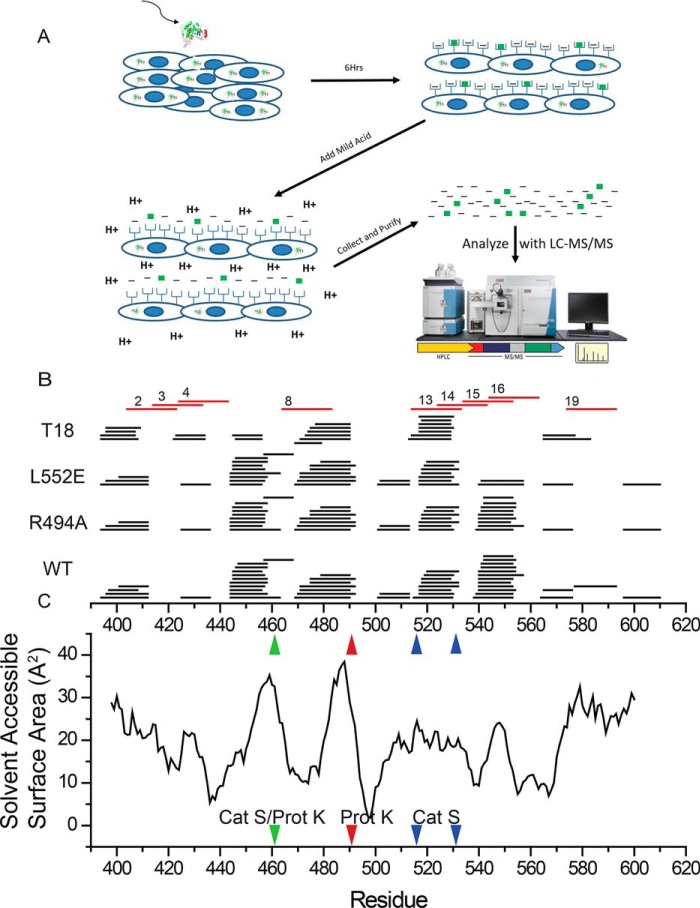
Figure 5.
In vitro processing and presentation of PE-III variants generate nested clusters of peptides. A, schematic for the mild acid elution workflow. Antigen-presenting cells are incubated with PE-III protein for 6 h before peptides are eluted with acid and analyzed by LC-MS/MS. B, eluted peptides detected by MS aligned to the PE-III sequence (black lines). Numbered red lines indicate PE-III epitopes reported here. C, solvent-accessible surface area determined from the WT PE-III structure (PDB code 1XK9) variant aligned to the graph in B. Red arrowhead indicates approximate locations of proteinase K cleavage site; blue arrowheads indicate approximate locations of cathepsin S cleavage sites, and the green arrowhead indicates a shared proteinase K (Prot K) and cathepsin S (Cat S) cleavage site.