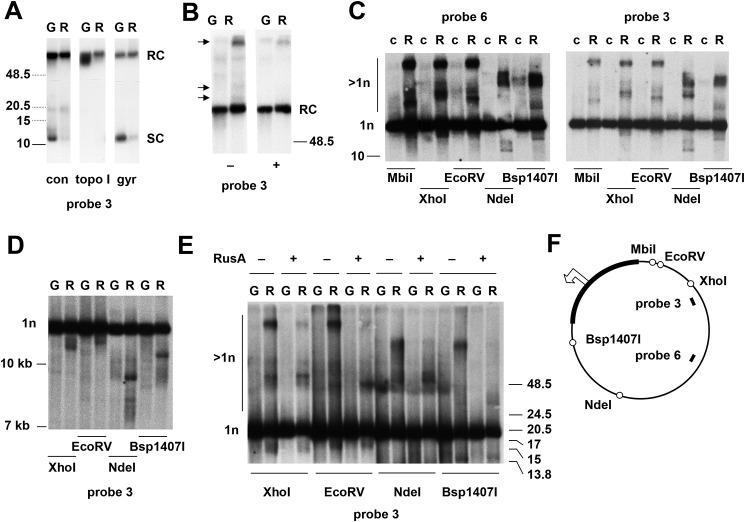
Figure 5.
rnh1 knockdown leads to the accumulation of abnormal mtDNA species. A–E, Southern blots of mtDNA (mitochondrial nucleic acid) from control S2 cells (c) or S2 cells treated with an inert dsRNA against GFP (G) or a dsRNA against rnh1 (R), probed as indicated. Migration of molecular weight markers (in kb) is indicated: dotted species in A are extrapolated from a separate gel of control material. A, undigested DNA, either untreated (con) or treated with topoisomerase I (topo I) or DNA gyrase (gyr). The forms inferred to be monomeric supercoiled circles (SC) or relaxed circles (RC) are as indicated. B, undigested DNA treated with (+) or without (−) S1 nuclease as shown. Arrowheads indicate molecular species unique to rnh1 knockdown cells that were sensitive to S1 nuclease. The forms inferred to be monomeric relaxed circles (RC) are as indicated. C and E, DNA digested with restriction enzymes as shown. The forms inferred to be monomeric linears (1n) or species of higher apparent molecular weight (>1n) are as indicated. Samples in E were treated with (+) or without (−) RusA as indicated. D, DNA digested with restriction enzymes as shown. F, schematic map of Drosophila mtDNA, indicating the location of relevant restriction sites (open circles), the noncoding region (bold), and the probes used. The open arrowhead marks the location and direction of replication initiation (see Ref. 40), near which one of the double-stranded ends inferred in D maps.