Abstract
The naturally occurring R68S substitution of phenylalanine hydroxylase (PheH) causes phenylketonuria (PKU). However, the molecular basis for how the R68S variant leads to PKU remains unclear. Kinetic characterization of R68S PheH establishes that the enzyme is fully active in the absence of allosteric binding of phenylalanine, in contrast to the WT enzyme. Analytical ultracentrifugation establishes that the isolated regulatory domain of R68S PheH is predominantly monomeric in the absence of phenylalanine and dimerizes in its presence, similar to the regulatory domain of the WT enzyme. Fluorescence and small-angle X-ray scattering analyses establish that the overall conformation of the resting form of R68S PheH is different from that of the WT enzyme. The data are consistent with the substitution disrupting the interface between the catalytic and regulatory domains of the enzyme, shifting the equilibrium between the resting and activated forms ∼200-fold, so that the resting form of R68S PheH is ∼70% in the activated conformation. However, R68S PheH loses activity 2 orders of magnitude more rapidly than the WT enzyme at 37 °C and is significantly more sensitive to proteolysis. We propose that, even though this substitution converts the enzyme to a constitutively active enzyme, it results in PKU because of the decrease in protein stability.
Keywords: allosteric regulation, small-angle X-ray scattering (SAXS), enzyme kinetics, analytical ultracentrifugation, hydroxylase, protein conformation, enzyme stability, phenylalanine hydroxylase, phenylketonuria, allostery, protein structure
Introduction
Phenylalanine hydroxylase (PheH)2 catalyzes the hydroxylation of phenylalanine in the liver to tyrosine using tetrahydrobiopterin (BH4) and molecular oxygen (1). The enzyme is tightly regulated to prevent accumulation of excess phenylalanine while maintaining the basal level of phenylalanine needed for cellular metabolism (2–4). The resting form of the enzyme is generally accepted to have low activity, becoming activated at high concentrations of phenylalanine when phenylalanine binds at an allosteric site (3, 5).
PheH is a homotetrameric enzyme containing three domains: an N-terminal regulatory domain with an ACT core (6), a larger catalytic domain (7), and a smaller C-terminal tetramerization domain (5). The structure of the intact resting form of the enzyme was described recently (8, 9), but there is as yet no high-resolution structure available for the activated form. These structures and an earlier structure of a dimeric form of the enzyme (10) show that the N terminus reaches across the active site. This region of the protein has been proposed to act as an autoinhibitory peptide, hindering binding of substrates to the active site (11, 12).
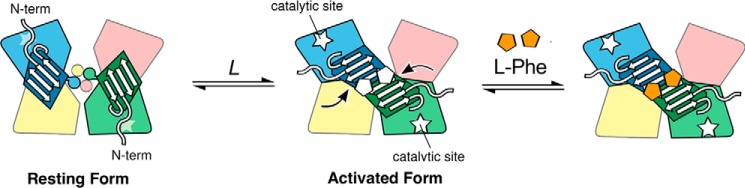
We have proposed a model for allosteric activation of PheH (Fig. 1) in which the regulatory domains form homodimers that can bind two phenylalanine molecules at the dimer interface; the movement of the regulatory domains to form dimers opens the active site by moving the autoinhibitory N-terminal residues away from the active site (8). This model is supported by small angle X-ray scattering (SAXS) analyses of the activated form (8), analytical ultracentrifugation analyses showing that phenylalanine stabilizes the regulatory domain dimer (13), analyses of the kinetic mechanism of activation (14), NMR experiments identifying the phenylalanine-binding residues at the dimer interface (15), and a crystal structure of the phenylalanine-bound regulatory domain dimer (16).
Figure 1.
Model for allosteric regulation of phenylalanine hydroxylase (8, 14). Catalytic domains are shown in blue, magenta, yellow, and green; the C-terminal helices are depicted as circles; and the regulatory domains corresponding to the blue and green catalytic domains are shown as trapezoids. The allosteric sites for phenylalanine at the interface of the regulatory domain dimer in the activated form are shown as pentagons.
Dysfunctional PheH leads to the metabolic disorder phenylketonuria (PKU), a disease that occurs in about 1 in 10,000 newborns in the United States (17). Over 1,000 PKU-causing mutations have been identified in PheH, with mechanisms of action ranging from misfolding to active site disruption (PAHvdb: phenylalanine hydroxylase gene locus-specific database; http://www.biopku.org/pah/home.asp).3 The effects on the enzyme of a number of PKU-causing mutations have been examined to understand the basis for their mechanism of action. Gjetting et al. (19) studied the effects of several PKU-causing mutations on the dimerization of regulatory domain fusion constructs and reported that the R68S substitution decreases the affinity of phenylalanine for the allosteric site (20), providing a potential explanation for why this mutation would cause PKU. However, activity studies of the intact enzyme suggested the opposite: Martínez and co-workers (21) reported that R68A PheH is fully active compared with the WT enzyme even in the absence of activation by phenylalanine. Taken together, these reports suggest that PheH R68S can be activated without phenylalanine binding in the allosteric site, possibly through a novel mechanism. However, it is not obvious why a constitutively active PheH would result in PKU. Herein, we characterize the PKU-causing substitution R68S to investigate the involvement of Arg-68 in the activation of PheH and the molecular basis for the contribution of this mutation to PKU.
Results
Effect of the R68S mutation on PheH steady-state kinetic parameters
In our hands, R68S PheH was readily expressed in Escherichia coli using the protocol developed for the WT enzyme. However, the enzyme precipitated when purified as described previously for WT PheH (8). Increasing the concentration of glycerol in purification buffers to 15–20% (v/v) significantly decreased the extent of precipitation, allowing the mutant protein to be purified.
We determined the steady-state kinetic parameters of R68S PheH for comparison with those for WT PheH. In contrast to the WT enzyme, pretreatment with phenylalanine does not increase the rate of phenylalanine hydroxylation by R68S PheH (Fig. S1 and Table 1). Instead, the resting form of the mutant enzyme is fully active. In addition, the R68S enzyme shows little or no cooperativity, unlike the WT enzyme.
Table 1.
Steady-state kinetic parameters of R68S and WT PheH
Conditions were as follows: 80 mm Hepes, pH 7.0, 50 μg/ml catalase, 1 mm dithiothreitol, 250 μm tetrahydrobiopterin, and 5 μm ferrous ammonium sulfate, pH 7.0, 23 °C. phe, phenylalanine.
| Enzyme | kcat | KPhe | KBH4a | nH |
|---|---|---|---|---|
| min−1 | μm | μm | ||
| R68S PheHa | 509 ± 22 | 221 ± 26 | 10.5 ± 0.2 | |
| R68S PheH plus 1 mm phea,b | 508 ± 22 | 212 ± 25 | 9.6 ± 3.4 | |
| R68S PheH without or plus 1 mm Phec,d | 466 ± 31 | 176 ± 29 | 10 ± 4 | 1.1 ± 0.1 |
| WT PheHd | 201 ± 10 | 231 ± 18 | 5.0 ± 2.3 | 2.4 ± 0.4 |
| WT Phe plus 1 mm Pheb,d | 353 ± 13 | 204 ± 13 | 13 ± 2 | 2.9 ± 0.5 |
a The data with phenylalanine as the varied substrate were fit to the Michaelis–Menten equation.
b Enzyme was pretreated with 1 mm phenylalanine at 23 °C for 3 min prior to starting the reaction.
c The data in the presence and absence of phenylalanine combined.
d The data with phenylalanine as the varied substrate were fit to the Hill equation. The values are from the combined data for 2–3 enzyme preparations in each case.
Fluorescence spectroscopy of R68S PheH and WT PheH
When activated by phenylalanine, the fluorescence emission of WT PheH increases, whereas the emission maximum shifts from ∼334 to ∼342 nm (Fig. 2A) (22). This is due to changes in the environment of Trp-120 (23), located in the interface between the catalytic and regulatory domains. The emission maximum of R68S PheH is at 342 nm, more consistent with that of the activated form than that of the resting form of WT PheH (Fig. 2B). There is no detectable change in the emission spectrum of R68S PheH in the presence of 1 mm (Fig. 2B) or 10 mm phenylalanine (data not shown). These results suggest that the environment of Trp-120 in the mutant protein differs from that in the resting form of WT PheH and are consistent with the conformation of the resting form of the mutant enzyme resembling that of the activated WT protein. The results are also consistent with the mutation resulting in a loss of binding of phenylalanine in the allosteric site.
Figure 2.
Fluorescence emission spectra of WT PheH (A) and R68S PheH (B) in the absence (dashed line) and presence (solid line) of 1 mm phenylalanine. All samples were excited at 295 nm at 23 °C, pH 7.5. The data for the WT enzyme are from Khan and Fitzpatrick (14).
Analytical ultracentrifugation of the isolated regulatory domain of R68S PheH
The isolated regulatory domain of PheH WT (RDPheH) exists in a monomer–dimer equilibrium, and the dimer is stabilized by the binding of phenylalanine at the dimer interface (13). To determine the effect of the R68S substitution on the oligomerization state of the regulatory domain, we carried out sedimentation velocity experiments. We used R68S RDPheH(25–117) for these experiments because this truncated form of RDPheH is better-behaved, and the loss of the unstructured N-terminal 24 residues has no effect on dimerization (13). Fig. 3 shows van Holde–Weischet analyses of sedimentation velocity experiments with R68S RDPheH(25–117) and WT RDPheH(25–117). The mutant regulatory domain has an average sw value very close to that of the WT enzyme, establishing that it is not constitutively dimeric in the absence of phenylalanine.
Figure 3.

Van Holde-Weischet analysis of R68S RDPheH(25–117). Shown are WT RDPheH(25–117) (black) and R68S RDPheH(25–117) (red) in the absence (solid circles) and presence (open circles) of 1 mm phenylalanine. The RDPheH(25–117) data are from Zhang et al. (13).
Analytical ultracentrifugation (AUC) analysis of the isolated regulatory domain also provides a direct measure of whether phenylalanine binds in the allosteric site. As shown in Fig. 3, the sw values of both the WT and the mutant enzymes increase in the presence of 1 mm phenylalanine to average values consistent with the formation of dimeric species. Thus, the mutant regulatory domain dimerizes in the presence of phenylalanine. The curves for the mutant domain suggest that R68S RDPheH consists of multiple species, including aggregates, in the presence and absence of phenylalanine. Due to this complication, the effect of the mutation on regulatory domain dimerization was not analyzed further. Still, these results establish that the regulatory domain R68S PheH by itself is predominantly monomeric in the absence of phenylalanine, forming a dimer in the presence of phenylalanine. This result rules out the possibility that the high activity of the resting form of the mutant protein is due to a constitutively dimerized regulatory domain and establishes that phenylalanine can still bind the allosteric site in the mutant protein.
SAXS
SAXS with in-line size-exclusion chromatography was used to determine the oligomeric state and probe changes in the domain configuration of full-length R68S PheH in solution. The protein eluted from the gel filtration column as a single narrow peak that could be separated computationally into individual components: 95% tetramer plus a small amount of aggregation and a dimer fraction of <5%. The scattering profile of the tetramer in the absence of phenylalanine has a radius of gyration of 40.9 ± 0.1 Å (Guinier, Fig. S2) or 40.6 Å (real-space) and a mid-q shape different from the curves for the resting and activated WT enzymes (Fig. 4A). The addition of 1 mm phenylalanine to R68S PheH results in a small decrease of the radius of gyration to 40.1 ± 0.1 Å (both methods) and a scattering profile that more strongly resembles that of the activated WT enzyme (Fig. 4B) (8). Fig. 5 shows the effects of increasing concentrations of phenylalanine on the scattering profile of 25 μm (monomer) R68S PheH. There is an isosbestic point at q ∼ 0.1 Å−1, consistent with a two-state equilibrium that can be analyzed by singular-value decomposition, as in our previous study of the WT PheH (8). The fit of the phenylalanine concentration dependence of the scattering profile to a noncooperative binding model (Equation 1) yields an apparent Kphe value of 29.6 ± 0.4 μm (Fig. 5, inset). Fitting to the Hill equation yields a Kphe value of 28.3 ± 3.7 μm and a Hill coefficient of 0.8 ± 0.1 (not shown). A similar titration for the WT enzyme showed cooperative binding of phenylalanine, with a Kphe value for phenylalanine of 174 μm and a Hill coefficient of 2.0 (8).
| (Eq. 1) |
Figure 4.
Kratky representations of the SEC-SAXS intensities of tetrameric PheH R68S and WT PheH in the absence (A) and in the presence (B) of phenylalanine. The WT data are from Meisburger et al. (8).
Figure 5.

Phenylalanine dependence of the SAXS profile of R68S PheH. Inset, change in the scattering curves of R68S PheH upon binding phenylalanine fit to Equation 1. a.u., arbitrary units.
Stability of R68S PheH
The stability of R68S PheH was determined by monitoring the enzyme activity upon incubation at pH 7 and 37 °C. Whereas the WT enzyme retains full activity after 3 h, the mutant enzyme loses one-half of its activity in 38 min under these conditions (Fig. 6).
Figure 6.

Stability of R68S (●) and WT PheH (□) at 37 °C. The enzymes were incubated at 37 °C in in 0.2 m Hepes, pH 7.0, and the remaining activity was determined at the indicated times. The data are from the averages of three separate experiments. The line is from a fit of the data to an exponential decay with an end point of zero. Error bars, S.D.
To further probe the effect of the R68S mutation on the protein structure, we carried out limited proteolysis experiments on WT and R68S PheH. R68S PheH is cleaved much more rapidly by trypsin than the WT protein, with almost complete cleavage of the 52-kDa protein to a fragment with a molecular mass of ∼38 kDa occurring after 1 h (Fig. 7 and Fig. S3). This is consistent with the loss of the regulatory domain, as was observed previously upon limited proteolysis of the WT enzyme (24). In the presence of phenylalanine, the WT and R68S PheH proteins are both protected from proteolysis (Fig. 7). This suggests that the domain configuration of R68S PheH in the presence of phenylalanine is similar to that of the activated WT PheH, in accord with the SAXS results. The extent of proteolysis of the mutant protein in the absence of phenylalanine differs from that of the WT protein with or without phenylalanine, suggesting that the domain configuration of R68S PheH in its resting state differs from that of the activated WT protein.
Figure 7.
SDS-PAGE of limited proteolysis by trypsin of WT and R68S PheH in the absence and presence of phenylalanine by trypsin. The enzymes (0.8 mg/ml) were incubated with 16 μg/ml trypsin at 23 °C and quenched at the indicated times with 1 mg/ml phenylmethylsulfonyl fluoride. When denoted, WT and R68S PheH samples were incubated with a final concentration of 1 mm phenylalanine for 3 min at 23 °C prior to the reaction.
Discussion
The association of the R68S substitution in PheH with PKU is counterintuitive in that the enzyme is fully active in the absence of allosteric activation. The results presented here allow us to rationalize this result within the framework of our mechanism for allosteric activation. They also provide additional insight into the molecular basis for regulation of PheH.
Our mechanism for the activation of PheH (8) involves a conformational change in which the regulatory domains dimerize, forming an allosteric phenylalanine site at the newly formed dimer interface (Fig. 1). This movement opens up the active site by displacing the N-terminal autoinhibitory (11) peptide. One possible explanation for the constitutive activity of the R68S enzyme is that the mutation stabilizes the regulatory domain dimer, so that the activated conformation is present even in the absence of phenylalanine. However, the AUC data show that the R68S regulatory domain undergoes a monomer–dimer equilibrium similar to that of the WT regulatory domain and that the addition of 1 mm phenylalanine stabilizes the dimer. These data establish that the resting form R68S PheH is not activated due to the presence of a constitutively dimerized regulatory domain; therefore, the structure of the resting form must be affected by the mutation.
The fluorescence, SAXS, and limited proteolysis results establish that the structure of the resting form of R68S PheH differs from that of both the resting and the activated WT enzyme. The changes in the intrinsic fluorescence of PheH mainly reflect the environment near Trp-120 (23), which is at the interface between the regulatory and catalytic domains in the resting conformation. The conformational change that accompanies activation of the WT enzyme by allosteric binding of phenylalanine in the regulatory domain necessarily involves movement of the regulatory domain away from the catalytic domain, allowing the regulatory domains to dimerize. This movement disrupts the protein structure in the vicinity of Trp-120, and the altered environment of this residue is reflected in the altered fluorescence of the activated protein. The agreement of the fluorescence emission maxima of the resting form of the mutant protein with that of the activated WT enzyme is consistent with the environment of Trp-120 resembling that of the latter, with the interface between the regulatory and catalytic domains being disrupted.
The different effects of limited proteolysis by trypsin on WT and R68S PheH similarly show that the resting forms of the two proteins have different structures. In contrast, the proteolysis patterns for the two proteins in the presence of phenylalanine are indistinguishable, consistent with phenylalanine binding in the allosteric site of the mutant protein stabilizing the same conformation as allosteric binding to the WT enzyme.
SAXS provides a probe of the global conformation of the mutant enzyme, and the scattering curves of the resting and phenylalanine-bound forms of R68S PheH are fully consistent with the fluorescence and proteolysis data. The mid-q region of the scattering curve is especially sensitive to the structural transition upon activation of PheH. In the absence of phenylalanine, the scattering profile of R68S PheH has a mid-q shape that is different from those of the resting and activated WT proteins. When phenylalanine is added, the scattering curve of the mutant protein changes to one that strongly resembles that of the activated WT PheH (Fig. 4B). The AUC results confirm that the R68S regulatory domains are able to form dimers under these conditions. The structure of the phenylalanine-bound isolated regulatory domain of PheH shows that Arg-68 is not involved in direct binding of phenylalanine (Fig. 8A) and does not lie in the dimer interface. Thus, there is no structural impediment in the mutant protein to dimerization of the regulatory domain or to the subsequent binding of phenylalanine at the allosteric site at the dimer interface, consistent with the conclusions from the AUC and SAXS results above.
Figure 8.

Location of Arg-68 in the PheH structure: Arg-68 (cyan) in the isolated regulatory domain with phenylalanine bound (green) (PDB code 5FII) (A) and Arg-68 in the resting form of intact PheH with adjacent subunits colored in blue and tan (PDB code 5EGQ) (B).
We considered two different models for the conformation of the resting form of R68S PheH. In both, the mutation disrupts the interface between the catalytic and regulatory domains. In the first, the regulatory domain has become undocked from the catalytic domain but has not formed a dimer. This would displace the regulatory domain from its position in the resting form of the WT enzyme, thereby altering the position of the autoinhibitory N terminus. In the second model, the mutation results in an altered equilibrium (L, Fig. 1) between the resting and activated conformations found in WT PheH.
To test the first hypothesis, we constructed a pool of 100 atomic models where the regulatory domains were undocked and connected to the catalytic domains only through a flexible linker (residues 110–118). The combination of models whose predicted scattering profile gave the best fit with the data is shown in Fig. 9A. The fit of the ensemble (curve II; Fig. 9B) is significantly better than the SAXS profile of the resting WT PheH (curve I). The chosen structures are not unique in the following sense: if a different pool were generated, one would expect to select a different set of structures and weights that agree similarly well with the data. Because the ensemble fit is not unique, and because SAXS is a low-information technique, we chose not to analyze in any detail the particular structures that were selected.
Figure 9.

Evaluation of atomic models for PheH R68S in the resting state. A, three candidate models for tetrameric R68S were evaluated: the crystallographic structure of the inactive WT PheH (PDB code 5FGJ) (I); a minimum ensemble selected from a pool of randomly generated structures with undocked regulatory domains (II); and a mixture of inactive and active WT conformations (III) (8). The catalytic and tetramerization domains are gray, and the regulatory domains are red. B, scattering curves predicted from models I, II, and III (solid lines, top panel) are compared with the SAXS profile of PheH R68S in the absence of phenylalanine (blue, gray, and magenta circles; top). The SAXS profile of PheH R68S in the presence of 1 mm phenylalanine is shown for reference (orange circles; top). Curves are offset for clarity. The residuals (bottom; corresponding colors) are largest for model I (χ2 = 7.4), intermediate for model II (χ2 = 5.6), and smallest for model III (χ2 = 2.8).
To test the second hypothesis, we fit the R68S SAXS profile to a linear combination of the scattering curves for models of the resting and activated WT enzymes (Fig. S4). A mixture of 67% activated conformation and 33% resting conformation (curve III) results in an excellent match to the profile for the resting form of the mutant enzyme. Fitting all of the curves in the phenylalanine titration shown in Fig. 5 to a linear combination of WT curves collected under similar conditions (8) successfully models those data and yields an L value of 1.4 ± 0.3 for R68S PheH (Fig. S4) compared with a value of 0.007 for the WT enzyme (8, 14). Thus, the SAXS results are consistent with a ∼200-fold shift in the equilibrium constant for formation of the activated conformation as a result of the mutation.
The loss of cooperativity in the mutant enzyme is compatible with a model in which L ∼ 1. As noted in the original description of the two-state model for allostery (30), an L value of ∼1 is effectively indistinguishable from a noncooperative system. An L value of 1.4 is also compatible with the fluorescence data. A model for the resting form of the mutant enzyme as a mixture containing 67% the activated conformation predicts a fluorescence maximum of ∼340 nm based on the 8-nm difference in the emission maxima of the resting and activated forms of the WT enzyme.
A value of L of 1.4 for the mutant protein makes the resting form of this enzyme a model for the intermediate in Fig. 1, because ∼70% of the protein is in this conformation in the absence of phenylalanine. There are no direct structural data available for this form of the WT enzyme. However, successful modeling of the kinetics of activation of PheH by phenylalanine requires the involvement of an intermediate with a fluorescence emission spectrum very similar to that of the activated WT enzyme (14); this is consistent with the fluorescence properties of the resting form of R68S PheH. The resting form of the mutant enzyme is significantly more sensitive to proteolysis than either the resting or activated forms of the WT enzyme. This may reflect a more dynamic structure in the absence of phenylalanine rather than a change in the overall structure. Finally, it should be noted that the R68S enzyme is a tetramer in the absence of phenylalanine, as is the WT enzyme in both the resting and activated conformation.
In the structure of the resting form of the intact WT enzyme, Arg-68 in the regulatory domain is in close proximity to the catalytic domain of the adjacent subunit. There is a network of interactions involving Arg-68 and Glu-66 in the regulatory domain of one subunit and Thr-236, Cys-237, and Arg-420 in the catalytic domain of another (Fig. 8B). Removal of the side chain of Arg-68 would be expected to disrupt this network and weaken the interaction between the regulatory and catalytic domains. Indeed, chemical modification of Cys-237, the thiol of which is in van der Waals contact with the δ-carbon of Arg-68, has an effect similar to that of the R68S mutation, generating an enzyme that is fully active without allosteric activation (25, 26). C237S PheH still requires activation by phenylalanine and shows a change in its fluorescence emission spectrum similar to that of the WT enzyme in response to phenylalanine (27), so that the activation upon chemical modification of this residue can be attributed to introduction of a bulky modifying reagent at this crucial interface. In addition, the E66K (28) and R420M (29) mutations in PheH both result in PKU.
PheH can also be converted to a fully active form in the absence of phenylalanine by incubation with lysolecithin (25, 32, 33). The mechanism by which this occurs is unknown. Treatment of PheH with lysolecithin also increases the reactivity of Cys-237 (33), consistent with binding of the phospholipid, resulting in a conformational change that exposes Cys-237 to solvent. All of these observations can be reconciled by a model in which modification of Cys-237, mutagenesis of Arg-68, or treatment with lysolecithin disrupt the interface between the catalytic and regulatory domains of PheH, shifting the value of L.
The altered regulatory properties of R68S PheH are unlikely to be responsible for the finding that this mutation is associated with PKU. Instead, the detrimental effects of this mutation can be attributed to its effect on the stability of the protein. With a half-life of under 1 h at 37 °C compared with the reported half-life of 2 days for the WT enzyme (34), the mutant protein loses activity almost 100 times more quickly. This would be expected to greatly decrease the steady-state level of the mutant protein in the liver, providing a reasonable explanation for why this mutation results in PKU. The instability of R68S PheH is also supported by the requirement of 20% glycerol in the storage buffer. The altered regulatory properties, with the resting form being fully active, could partially ameliorate the decrease in stability. A similar decrease in stability probably accounts for the association of the E66K and R420M mutations with PKU.
The present results for R68S PheH support the hypothesis that the substitution disrupts an interaction between the regulatory domains and catalytic domains of adjacent subunits needed for the inhibited conformation of the resting WT enzyme. Disruption of these interactions results in activated enzyme even in the absence of phenylalanine binding at the allosteric site but also makes the protein significantly less stable.
Experimental procedures
Materials
BL21(DE3) cells were from Stratagene Corp. (La Jolla, CA). The pGro7 plasmid was from Takara Bio, Inc. (Otsu, Shiga, Japan). Pfu polymerase was from Agilent (Santa Clara, CA). Ampicillin was purchased from Research Products International Corp. (Mount Prospect, IL). Chloramphenicol was from Acros Organics (Geel, Belgium). l-Arabinose was from Alfa Aesar (Ward Hill, MA). Tetrahydrobiopterin was from Schircks Laboratories (Bauma, Switzerland). Tween 20 was from EMD Millipore Corp. (Billerica, MA). Isopropyl β-d-1-thiogalactopyranoside was from Gold Biotechnology, Inc. (St. Louis, MO). Leupeptin was from Peptide Institute, Inc. (Ibaraki-Shi, Osaka, Japan). Lysozyme was from MP Biomedicals (Santa Ana, CA). Other reagents were purchased from Sigma-Aldrich or Fisher. The plasmids for expression of rat PheH (pERPH2.0) and the regulatory domain of PheH lacking the first N-terminal 24 residues (pEThisRD(25–117)) were described previously (12, 35). The R68S mutation was introduced into both using the QuikChange (Agilent) site-directed mutagenesis method with oligonucleotides from Integrated DNA Technologies. E. coli BL21(DE3) cells containing plasmid pGro7 (Takara) were transformed with the resulting construct (R68S pERPH2.0 or pETR68ShisRD). The coding regions of all expression plasmids were sequenced by GenScript Biotech Corp. to verify the mutation and to ensure no other mutations were present.
Protein expression and purification
R68S PheH was expressed and purified as described previously for WT PheH with the following modifications (12). The phenyl-Sepharose column was washed with lysis buffer (30 mm Hepes, 0.2 m NaCl, 20 mm phenylalanine, 200 μm diethylenetriaminepentaacetic acid, 1 μm leupeptin, and 1 μm pepstatin, pH 7.2) containing 1 mm ATP and 1 mm MgCl2 to elute the GroEL/ES chaperone immediately prior to R68S PheH elution with 30 mm Hepes, 20% glycerol, 50 μm diethylenetriaminepentaacetic acid, 1 μm leupeptin, and 1 μm pepstatin, pH 7.2. The PheH-containing fractions were applied to a DEAE column and eluted with 50 mm Hepes, 15% glycerol, pH 7.0, with a gradient of 0–500 mm NaCl in the same buffer. The fractions with the highest purity were then applied to a HiLoad 16/60 Superdex 200 preparation grade column (GE Healthcare) equilibrated with 50 mm Hepes, 20% glycerol, pH 7.0. The resulting fractions were analyzed by SDS-PAGE, and those containing >90% pure protein were pooled, concentrated to 250–300 μm, and stored at −80 °C. R68S PheH(25–117) was expressed and purified as described previously for the WT regulatory domain (13). Protein concentrations were determined using the extinction coefficients at 280 nm (36).
Assays
PheH activity was determined by following the formation of tyrosine at 275 nm at 23 °C as described previously (36). The assay contained 80 mm Hepes, pH 7.0, 50 μg/ml catalase, 1 mm DTT, and 5 μm ferrous ammonium sulfate. When the concentration of phenylalanine was varied (10–1000 μm), the concentration of BH4 was fixed at 250 μm. When the pterin concentration was varied (6.25–200 μm), the phenylalanine concentration was fixed at 1 mm. Reactions were initiated with enzyme at a final concentration of 0.5 μm, and the reaction was monitored for 150 s. The initial rates v as a function of substrate concentration were fit with the Michaelis–Menten equation for BH4 or the Hill equation (Equation 2) for phenylalanine.
| (Eq. 2) |
Fluorescence spectroscopy
Fluorescence spectra were obtained on a Shimadzu RF-5301PC spectrofluorometer at 23 °C. A 3-ml sample of 1 μm protein was excited at 295 nm, and the emission was scanned from 300 to 450 nm. When the scan was repeated in the presence of 1 mm phenylalanine, the protein was incubated with 1 mm phenylalanine for 5 min at room temperature prior to the scan. Raman scattering was corrected by subtraction of the fluorescence intensity of buffer alone (0.2 m Hepes, pH 7.5).
AUC
The sedimentation velocity AUC experiments were performed as described previously (13) in the Center for Macromolecular Interactions at the University of Texas Health Science Center at San Antonio. A Beckman Coulter XL-I ultracentrifuge (Beckman Instruments) with an An60Ti four-hole rotor was used at 50,000 rpm at 20 °C. All samples were prepared in 100 mm sodium phosphate, 20% glycerol, pH 8.0. Protein concentrations were monitored at 230 nm. Van Holde–Weischet analyses were performed using Ultrascan III (37).
Limited proteolysis
Limited proteolysis experiments were performed as described previously (38) with modifications. Protein samples (0.8 mg/ml) were incubated with 16 μg/ml trypsin at 23 °C. Samples with phenylalanine were incubated with a final concentration of 1 mm phenylalanine for 3 min at 23 °C prior to the reaction. Aliquots (10 μl) were removed and quenched at various times with 2 μl of 1 mg/ml phenylmethylsulfonyl fluoride. The samples were mixed with SDS-PAGE sample buffer containing β-mercaptoethanol and applied to a 10% SDS-polyacrylamide gel.
Thermal inactivation
The R68S and WT PheH enzymes (0.5 μm) were incubated at 37 °C in 0.2 m Hepes, pH 7.0. Aliquots were removed at 0–180 min, and the tyrosine formation activity was assayed as described above with 300 μm phenylalanine and 200 μm BH4.
Chromatography-coupled small angle X-ray scattering
X-ray scattering measurements were performed at the G1 Station of the Cornell High Energy Synchrotron Source using a size-exclusion column in-line with the SAXS flow cell, as described previously (8, 39). The column (Superdex 10/300 Increase column, GE Healthcare Life Sciences) was pre-equilibrated with 50 mm Hepes, 20% glycerol, pH 7.0. Samples (100 μl of 260 μm R68S PheH) were injected into a 500-μl loop and passed through the column at 0.5 ml/min. Experiments in the presence of phenylalanine were performed similarly, except the running buffer contained 1 mm phenylalanine, and the sample was incubated with 1 mm phenylalanine at 25 °C for 5 min and centrifuged for 1 min prior to injection. The FPLC and SAXS cell were at 25 °C. SAXS data were collected as 450 exposures of 5 s each. The X-ray wavelength was 1.2493 Å, and the sample–detector distances were 1.5336 m (SAXS) and 0.4639 m (WAXS), for a combined q-range of 0.01–0.6 Å−1. The data sets were analyzed using evolving factor analysis (8) and decomposed by regularized alternating least squares (40) to obtain scattering curves for the tetrameric components. Phenylalanine titration experiments were performed with 25 μm R68S PheH with varying concentrations of phenylalanine (0–1 mm). Singular-value decomposition was performed as described previously (8), and the first right singular vector was fit with Equation 1, where ΔS is the change in the scattering curve, [phe] is the concentration of phenylalanine, Smax is the value at which all subunits are in the activated conformation, and Kphe is the apparent dissociation constant for phenylalanine binding. A pool of 100 atomic models with undocked regulatory domains was generated using RANCH (41). Domain coordinates were those of the WT PheH in the resting conformation (PDB entry 5FGJ) except residues 1–33 and 110–118, which were modeled as random coils. Missing atoms were added using Modeller (42), and the scattering patterns were simulated using CRYSOL (31) with solvent electron density of 0.35 Å−3 corresponding to the experimental buffer, which contained 20% glycerol (18). The best-fit ensemble was found using lsqnonneg in MATLAB (MathWorks, Natick, MA).
Author contributions
C. A. K., S. P. M., N. A., and P. F. F. conceptualization; C. A. K., S. P. M., N. A., and P. F. F. formal analysis; C. A. K. and S. P. M. investigation; C. A. K. methodology; C. A. K. and S. P. M. writing-original draft; C. A. K., S. P. M., N. A., and P. F. F. writing-review and editing; N. A. and P. F. F. supervision; P. F. F. project administration.
Supplementary Material
Acknowledgments
We thank Dr. Gregory Reinhart for helpful discussion during this work. Support for the Center for Macromolecular Interactions from the Office of the Vice President for Research at UT Health San Antonio is gratefully acknowledged. CHESS is supported by the National Science Foundation (NSF) and National Institutes of Health (NIH)/NIGMS via NSF Grant DMR-1332208, and the MacCHESS resource is supported by NIH/NIGMS Grant GM-103485.
Note added in proof
An incorrect version of Fig. 9 was inadvertently uploaded in the version of this article that was published as a Paper in Press on January 23, 2019. This error has now been corrected.
This work was supported in part by National Institutes of Health Grants R01GM098140 (to P. F. F.), F31GM116452 (to C. A. K.), F32GM117757 (to S. P. M.), and R35GM124847 (to N. A) and Welch Foundation Grant AQ-1245 (to P. F. F.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- PheH
- phenylalanine hydroxylase
- BH4
- tetrahydrobiopterin
- AUC
- analytical ultracentrifugation
- SAXS and WAXS
- small- and wide-angle X-ray scattering, respectively
- RDPheH
- the isolated regulatory domain of PheH
- R68S RDPheH(25–117)
- the isolated regulatory domain lacking the N-terminal 24 residues
- PKU
- phenylketonuria
- PDB
- Protein Data Bank.
References
- 1. Fitzpatrick P. F. (2003) Mechanism of aromatic amino acid hydroxylation. Biochemistry 42, 14083–14091 10.1021/bi035656u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Shiman R., Jones S. H., and Gray D. W. (1990) Mechanism of phenylalanine regulation of phenylalanine hydroxylase. J. Biol. Chem. 265, 11633–11642 [PubMed] [Google Scholar]
- 3. Fitzpatrick P. F. (2012) Allosteric regulation of phenylalanine hydroxylase. Arch. Biochem. Biophys. 519, 194–201 10.1016/j.abb.2011.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Flydal M. I., and Martinez A. (2013) Phenylalanine hydroxylase: function, structure, and regulation. IUBMB Life 65, 341–349 10.1002/iub.1150 [DOI] [PubMed] [Google Scholar]
- 5. Fitzpatrick P. F. (2015) Structural insights into the regulation of aromatic amino acid hydroxylation. Curr. Opin. Struct. Biol. 35, 1–6 10.1016/j.sbi.2015.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lang E. J., Cross P. J., Mittelstädt G., Jameson G. B., and Parker E. J. (2014) Allosteric ACTion: the varied ACT domains regulating enzymes of amino-acid metabolism. Curr. Opin. Struct. Biol. 29, 102–111 10.1016/j.sbi.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 7. Daubner S. C., Hillas P. J., and Fitzpatrick P. F. (1997) Expression and characterization of the catalytic domain of human phenylalanine hydroxylase. Arch. Biochem. Biophys. 348, 295–302 10.1006/abbi.1997.0435 [DOI] [PubMed] [Google Scholar]
- 8. Meisburger S. P., Taylor A. B., Khan C. A., Zhang S., Fitzpatrick P. F., and Ando N. (2016) Domain movements upon activation of phenylalanine hydroxylase characterized by crystallography and chromatography-coupled small-angle X-ray scattering. J. Am. Chem. Soc. 138, 6506–6516 10.1021/jacs.6b01563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arturo E. C., Gupta K., Héroux A., Stith L., Cross P. J., Parker E. J., Loll P. J., and Jaffe E. K. (2016) First structure of full-length mammalian phenylalanine hydroxylase reveals the architecture of an autoinhibited tetramer. Proc. Natl. Acad. Sci. U.S.A. 113, 2394–2399 10.1073/pnas.1516967113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kobe B., Jennings I. G., House C. M., Michell B. J., Goodwill K. E., Santarsiero B. D., Stevens R. C., Cotton R. G. H., and Kemp B. E. (1999) Structural basis of autoregulation of phenylalanine hydroxylase. Nat. Struct. Biol. 6, 442–448 10.1038/8247 [DOI] [PubMed] [Google Scholar]
- 11. Jennings I. G., Teh T., and Kobe B. (2001) Essential role of the N-terminal autoregulatory sequence in the regulation of phenylalanine hydroxylase. FEBS Lett. 488, 196–200 10.1016/S0014-5793(00)02426-1 [DOI] [PubMed] [Google Scholar]
- 12. Roberts K. M., Khan C. A., Hinck C. S., and Fitzpatrick P. F. (2014) Activation of phenylalanine hydroxylase by phenylalanine does not require binding in the active site. Biochemistry 53, 7846–7853 10.1021/bi501183x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhang S., Roberts K. M., and Fitzpatrick P. F. (2014) Phenylalanine binding is linked to dimerization of the regulatory domain of phenylalanine hydroxylase. Biochemistry 53, 6625–6627 10.1021/bi501109s [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khan C. A., and Fitzpatrick P. F. (2018) Phosphorylation of phenylalanine hydroxylase increases the rate constant for formation of the activated conformation of the enzyme. Biochemistry 57, 6274–6277 10.1021/acs.biochem.8b00919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang S., and Fitzpatrick P. F. (2016) Identification of the allosteric site for phenylalanine in rat phenylalanine hydroxylase. J. Biol. Chem. 291, 7418–7425 10.1074/jbc.M115.709998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Patel D., Kopec J., Fitzpatrick F., McCorvie T. J., and Yue W. W. (2016) Structural basis for ligand-dependent dimerization of phenylalanine hydroxylase regulatory domain. Sci. Rep. 6, 23748 10.1038/srep23748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Williams R. A., Mamotte C. D. S., and Burnett J. R. (2008) Phenylketonuria: an inborn error of phenylalanine metabolism. Clin. Biochem. Rev. 29, 31–41 [PMC free article] [PubMed] [Google Scholar]
- 18. Tyree T. J., Dan R., and Thorne R. E. (2018) Density and electron density of aqueous cryoprotectant solutions at cryogenic temperatures for optimized cryoprotection and diffraction contrast. Acta Crystallogr. D Struct. Biol. 74, 471–479 10.1107/S2059798318003078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gjetting T., Petersen M., Guldberg P., and Güttler F. (2001) Missense mutations in the N-terminal domain of human phenylalanine hydroxylase interfere with binding of regulatory phenylalanine. Am. J. Hum. Genet. 68, 1353–1360 10.1086/320604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisensmith R. C., and Woo S. L. (1992) Molecular basis of phenylketonuria and related hyperphenylalaninemias: mutations and polymorphisms in the human phenylalanine hydroxylase gene. Hum. Mutat. 1, 13–23 10.1002/humu.1380010104 [DOI] [PubMed] [Google Scholar]
- 21. Thórólfsson M., Teigen K., and Martínez A. (2003) Activation of phenylalanine hydroxylase: effect of substitutions at Arg68 and Cys237. Biochemistry 42, 3419–3428 10.1021/bi034021s [DOI] [PubMed] [Google Scholar]
- 22. Phillips R. S., and Kaufman S. (1984) Ligand effects on the phosphorylation state of hepatic phenylalanine hydroxylase. J. Biol. Chem. 259, 2474–2479 [PubMed] [Google Scholar]
- 23. Knappskog P. M., and Haavik J. (1995) Tryptophan fluorescence of human phenylalanine hydroxylase produced in Escherichia coli. Biochemistry 34, 11790–11799 10.1021/bi00037a017 [DOI] [PubMed] [Google Scholar]
- 24. Abita J.-P., Parniak M., and Kaufman S. (1984) The activation of rat liver phenylalanine hydroxylase by limited proteolysis, lysolecithin, and tocopherol phosphate: changes in conformation and catalytic properties. J. Biol. Chem. 259, 14560–14566 [PubMed] [Google Scholar]
- 25. Parniak M. A., and Kaufman S. (1981) Rat liver phenylalanine hydroxylase: activation by sulfhydryl modification. J. Biol. Chem. 256, 6876–6882 [PubMed] [Google Scholar]
- 26. Gibbs B. S., and Benkovic S. J. (1991) Affinity labeling of the active site and the reactive sulfhydryl associated with activation of rat liver phenylalanine hydroxylase. Biochemistry 30, 6795–6802 10.1021/bi00241a024 [DOI] [PubMed] [Google Scholar]
- 27. Knappskog P. M., and Martínez A. (1997) Effect of mutations at Cys237 on the activation state and activity of human phenylalanine hydroxylase. FEBS Lett. 409, 7–11 10.1016/S0014-5793(97)00465-1 [DOI] [PubMed] [Google Scholar]
- 28. Bueno M. A., Lage S., Delgado C., Andrade F., Couce M. L., González-Lamuño D., Pérez M., and Aldámiz-Echevarría L. (2012) New evidence for assessing tetrahydrobiopterin (BH4) responsiveness. Metabolism 61, 1809–1816 10.1016/j.metabol.2012.07.015 [DOI] [PubMed] [Google Scholar]
- 29. Couce M. L., Bóveda M. D., Fernández-Marmiesse A., Mirás A., Pérez B., Desviat L. R., and Fraga J. M. (2013) Molecular epidemiology and BH4-responsiveness in patients with phenylalanine hydroxylase deficiency from Galicia region of Spain. Gene 521, 100–104 10.1016/j.gene.2013.03.004 [DOI] [PubMed] [Google Scholar]
- 30. Monod J., Wyman J., and Changeux J.-P. (1965) On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 12, 88–118 10.1016/S0022-2836(65)80285-6 [DOI] [PubMed] [Google Scholar]
- 31. Svergun D., Barberato C., and Koch M. H. J. (1995) CRYSOL: a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 28, 768–773 10.1107/S0021889895007047 [DOI] [Google Scholar]
- 32. Fisher D. B., and Kaufman S. (1972) The stimulation of rat liver phenylalanine hydroxylase by phospholipids. J. Biol. Chem. 247, 2250–2252 [PubMed] [Google Scholar]
- 33. Fisher D. B., and Kaufman S. (1973) The stimulation of rat liver phenylalanine hydroxylase by lysolecithin and α-chymotrypsin. J. Biol. Chem. 248, 4345–4353 [PubMed] [Google Scholar]
- 34. Chang N., Kaufman S., and Milstien S. (1979) The mechanism of the irreversible inhibition of rat liver phenylalanine hydroxylase due to treatment with p-chlorophenylalanine: the lack of effect on turnover of phenylalanine hydroxylase. J. Biol. Chem. 254, 2665–2668 [PubMed] [Google Scholar]
- 35. Li J., Ilangovan U., Daubner S. C., Hinck A. P., and Fitzpatrick P. F. (2011) Direct evidence for a phenylalanine site in the regulatory domain of phenylalanine hydroxylase. Arch. Biochem. Biophys. 505, 250–255 10.1016/j.abb.2010.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Daubner S. C., Hillas P. J., and Fitzpatrick P. F. (1997) Characterization of chimeric pterin-dependent hydroxylases: contributions of the regulatory domains of tyrosine and phenylalanine hydroxylase to substrate specificity. Biochemistry 36, 11574–11582 10.1021/bi9711137 [DOI] [PubMed] [Google Scholar]
- 37. Demeler B. (2005) UltraScan: a comprehensive data analysis software package for analytical ultracentrifugation experiments. in Modern Analytical Ultracentrifugation: Techniques and Methods (Scott D. J., Harding S. E., and Row A. J., eds) pp. 210–229, Royal Society of Chemistry, London [Google Scholar]
- 38. McCulloch R. I., and Fitzpatrick P. F. (1999) Limited proteolysis of tyrosine hydroxylase identifies residues 33–50 as conformationally sensitive to phosphorylation state and dopamine binding. Arch. Biochem. Biophys. 367, 143–145 10.1006/abbi.1999.1259 [DOI] [PubMed] [Google Scholar]
- 39. Acerbo A. S., Cook M. J., and Gillilan R. E. (2015) Upgrade of MacCHESS facility for X-ray scattering of biological macromolecules in solution. J. Synchrotron Radiat. 22, 180–186 10.1107/S1600577514020360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parker M. J., Maggiolo A. O., Thomas W. C., Kim A., Meisburger S. P., Ando N., Boal A. K., and Stubbe J. (2018) An endogenous dAMP ligand in Bacillus subtilis class Ib RNR promotes assembly of a noncanonical dimer for regulation by dATP. Proc. Natl. Acad. Sci. U.S.A. 115, E4594–E4603 10.1073/pnas.1800356115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Petoukhov M. V., Franke D., Shkumatov A. V., Tria G., Kikhney A. G., Gajda M., Gorba C., Mertens H. D. T., Konarev P. V., and Svergun D. I. (2012) New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Crystallogr. 45, 342–350 10.1107/S0021889812007662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fiser A., Do R. K. G., and Šali A. (2000) Modeling of loops in protein structures. Protein Sci. 9, 1753–1773 10.1110/ps.9.9.1753 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.