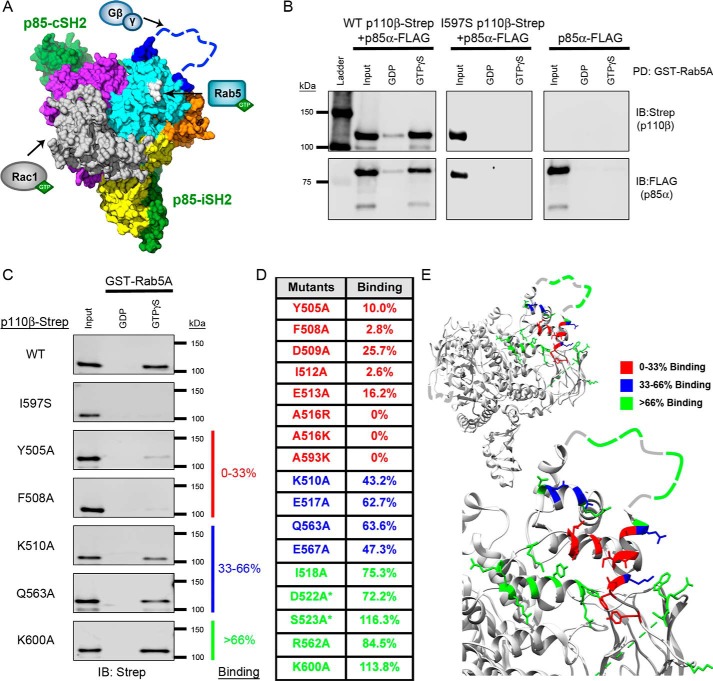
Figure 1.
Mutagenesis of the p110β helical domain disrupts Rab5A binding to PI3Kβ. A, space-filling model of the murine p110β catalytic subunit with the iSH2 and cSH2 domains of the p85β regulatory subunit (green) (Protein Data Bank (PDB) code 2Y3A). Yellow, the N-terminal ABD; gray, the RBD; orange, the C2 domain; cyan, the helical domain; purple, the C-terminal kinase domain. The blue dashed line represents the C2–helical linker, which was not observed in the X-ray structure. The arrows indicate where Rac1, Rab5, and Gβγ bind to p110β. Gln596/Ile597, whose mutation disrupts Rab5 binding, are shown in white. B, representative immunoblots (IB) of the GST-Rab5A pulldown (PD) assay. Human GST-Rab5A was immobilized on GSH-agarose beads and loaded with GDP or GTPγS. The beads were incubated with whole-cell lysates (Input) from HEK293T cells transfected with p85α-FLAG without or with WT p110β-Strep or the Rab5-uncoupled I597S mutant. C, representative immunoblots of the GST-Rab5A pulldown assay with p110β mutants. Samples were analyzed by SDS-PAGE and blotted for Strep (p110β) and FLAG (p85α). D, table of Rab5A-binding activity for representative p110β mutants. Binding to GTPγS–Rab5A was calculated as a percentage of the input and then normalized to WT p110β binding, which was set to 100%. Binding, as compared with WT p110β, was stratified into three groups: 0–33% (red), 33–66% (blue), and >66% (green). Residues in the Gβγ-binding loop are indicated with an asterisk. E, ribbon diagrams of p110β (upper panel) and a magnified view of the helical domain (lower panel). Residues are color-coded based on their Rab5A-binding activity (PBD code 2Y3A).