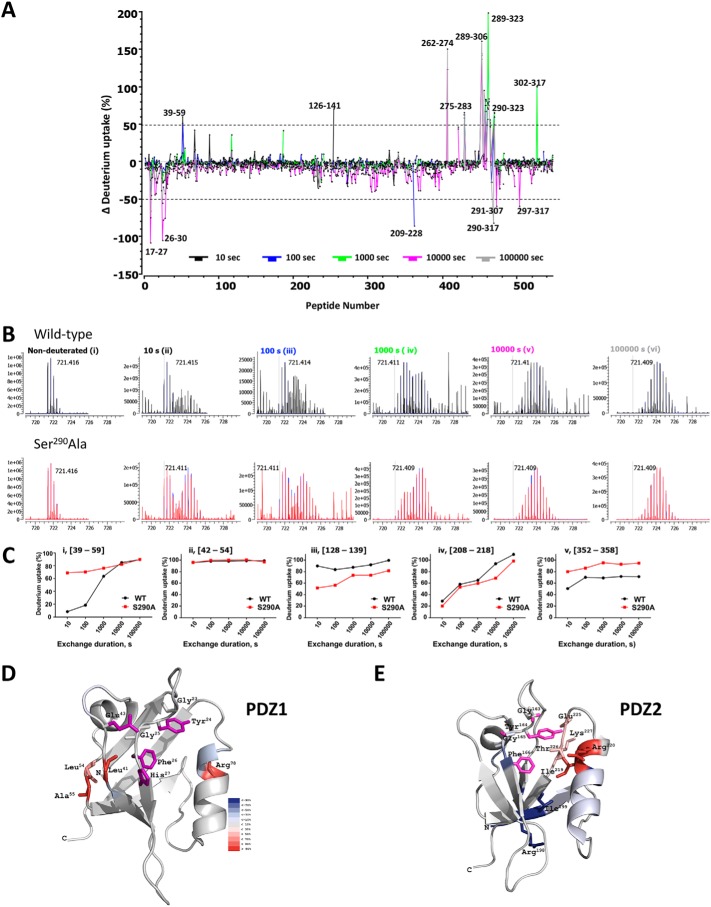
Figure 9.
Quantitative HDX analysis and NHERF1 conformation. A, differential deuterium uptake for WT–NHERF1 and S290A–NHERF1 and at 10 s (black), 100 s (blue), 1000 s (green), 10000 s (pink), and 100,000 s (gray). Peptides exhibiting significant differential deuterium uptake greater than 50% are indicated; others were omitted for clarity. Of note, there are large peaks of differential deuterium uptake surrounding Ser290 from Ala289 to Phe323 in the S290A phosphoresistant construct. The (39–58)-peptide, where Glu43 is adjacent to the PDZ1 23GYGF26 core-binding motif as shown in Fig. 8A, displays high differential deuterium uptake. B, representative isotope profile of the (39–58)-peptide at the indicated times for WT (black) and S290A–NHERF1 (red). Nondeuterated controls for WT and S290A peptides are indistinguishable. A bimodal distribution at 10, 100, and 1000 s (panels ii–iv) reflects conformational change between phosphorylated and unphosphorylated conditions. C, dynamic exchange profiles for the indicated peptides from WT (black) and S290A–NHERF1 (red). Peptides with unique, time-dependent HDX behavior are shown. C-terminal residues in S290A–NHERF1 display greater deuterium uptake. D, model showing residues exhibiting differential deuterium uptake between WT and S290A–NHERF1 at 100 s mapped on the structure of PDZ1 (PDB code 1I92). The critical residues His27, Glu43, and the GYGF core-binding motif for PDZ ligand binding are colored in purple. Residues undergoing increased deuteration of 30% or more are shown. E, differential deuterium uptake between WT and S290A–NHERF1 at 100 s mapped on PDZ2 (PDB code 4Q3H). Critical binding residues based on the same criteria as in D are colored purple. The differential deuteration levels are color-coded as in D. The high HDX rate at Ile219, Arg220, Glu225, Thr226, and Lys227 suggests that these residues undergo significant structural movement in the opposite direction from what occurs in PDZ1.