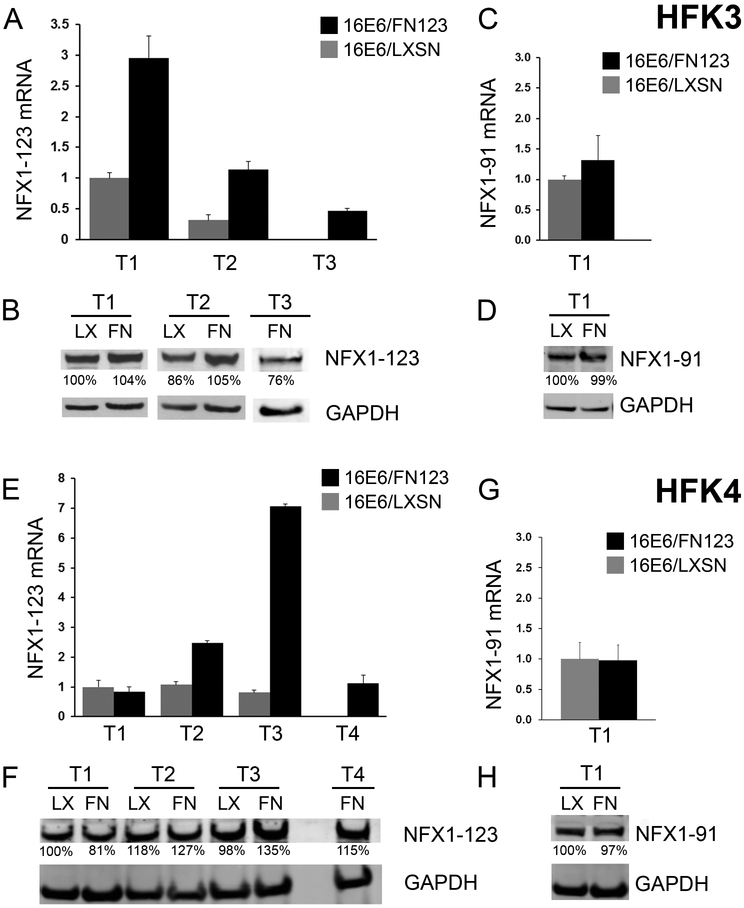
Figure 3. NFX1-123 overexpression was sustained in 16E6/FN123 HFKs.
(A-D) HFK3; T1 and T2 are matched shared timepoints for 16E6/LXSN and 16E6/FN123. T3 is one timepoint for 16E6/FN123 past the end of active growth for 16E6/LXSN (noted as vertical arrows in Figure 2C). (E-H) HFK4; T1-3 are matched shared timepoints for 16E6/LXSN and 16E6/FN123. T4 is one timepoint for 16E6/FN123 past the end of active growth for 16E6/LXSN (noted as vertical arrows in Figure 2D). (A) Mean expression of NFX1-123 mRNA in HFK3 at T1-T3, relative to 16E6/LXSN HFK3 at T1, was quantified. (B) NFX1-123 protein was increased in 16E6/FN123 HFK3 (FN) relative to the 16E6/LXSN HFK3 (LX). (C) Mean expression of NFX1-91 mRNA, relative to 16E6/LXSN HFK3 at T1, was quantified. (D) NFX1-91 protein was equivalent in 16E6/FN123 HFK3 compared to 16E6/LXSN. (E) Mean expression of NFX1-123 mRNA in HFK4 at T1-T4, relative to 16E6/LXSN HFK4 at T1, was quantified. (F) NFX1-123 protein was moderately increased in 16E6/FN123 HFK4 (FN) relative to the 16E6/LXSN HFK4 (LX). (G) Mean expression of NFX1-91 mRNA, relative to 16E6/LXSN HFK4 at T1, was quantified. (H) NFX1-91 protein was equivalent in 16E6/FN123 HFK4 compared to 16E6/LXSN. (A, C, E, and G: all qPCRs were normalized to the housekeeping gene 36B4, and all error bars represent 95% confidence intervals from the technical replicates shown (n = 3). B, D, F and H: GAPDH used as a loading control.)