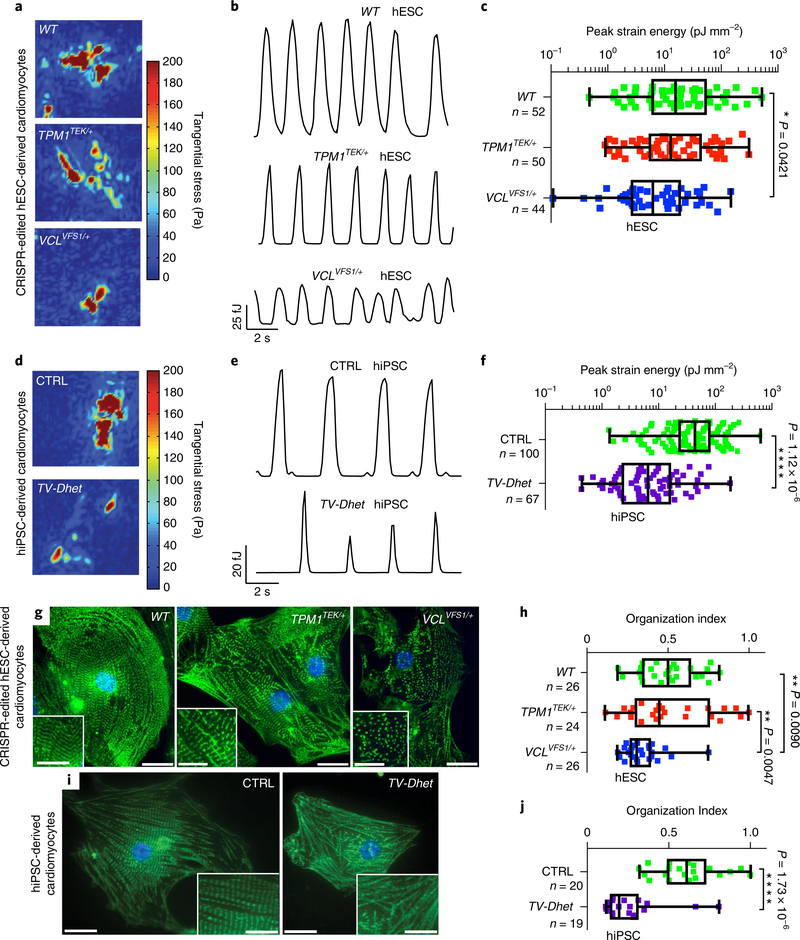
Fig. 2 |. hPSC-derived cardiomyocytes harbouring TEK; VCL genetic variants exhibit functional and sarcomeric organization defects.
a–f, Single-cardiomyocyte traction force microscopy studies show that TV-Dhet and VCLVFS1/+ hPSC-derived cardiomyocytes exhibit reduced contractility as detected by tangential stress heat maps (a,d), strain energy time courses (b,e) and peak strain energy analyses (c,f). g–j, Alpha-actinin immunostaining (green; g,i) and quantitation of the sarcomeric organization (h,j) reveal that sarcomeres of TV-Dhet and VCLVFS1/+ hPSC-derived cardiomyocytes are more disorganized than that of WT and CTRL cardiomyocytes. Control hESC-derived (WT) and CRISPR-edited-hESC-derived cardiomyocytes are represented in a–c,g,h. Control hiPSC-derived (CTRL) and cardiomyopathy-affected patient (TV-Dhet) hiPSC-derived cardiomyocytes are represented in d–f,i,j. Data are presented as interquartile range (box) around the mean (horizontal line), whiskers represent the minimum and maximum of the dataset, with the individual data points superimposed. *P < 0.05, **P < 0.01, ***P < 0.001 and ****P < 0.0001; two-sided Student’s t-test (f,j) or one-sided ANOVA with Tukey multiple correction test alpha (c,h). g,i, The representative images shown were obtained from experiments independently repeated three times with similar results; Hoechst nuclear labelling is shown in blue. Scale bars, 20 μm. Inset scale bars, 10 μm. n, number of biologically independent cardiomyocytes of each genotype used for each study.