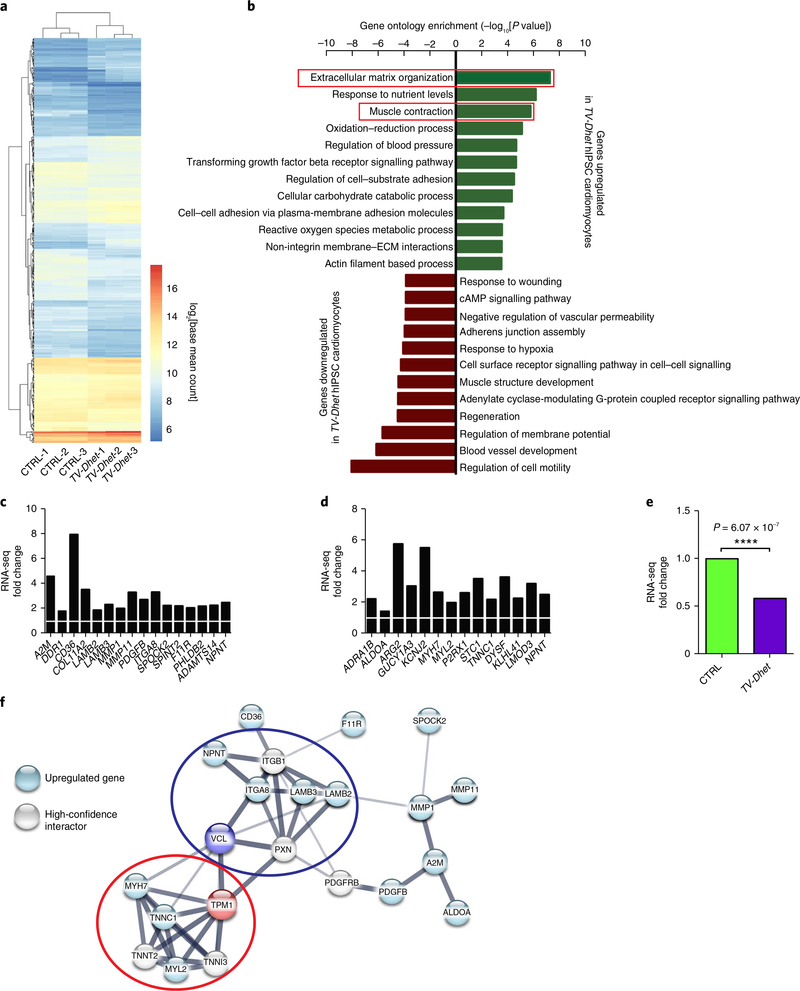
Fig. 3 |. ECM and muscle contraction genes are coordinately upregulated in TV-Dhet hiPSC cardiomyocytes.
a, Hierarchical clustering of differentially expressed genes between control (CTRL) and cardiomyopathy-affected patient (TV-Dhet) hiPSC-derived cardiomyocytes (n = 3 biological replicates per genotype) reveals clusters of genes that are coordinately regulated in TV-Dhet cardiomyocytes. Gene expression data from these replicates were combined by genotype in b–f to further compare relative gene expression changes. b, ECM organization and muscle contraction gene ontology terms are significantly enriched among genes upregulated in TV-Dhet cardiomyocytes whereas various gene ontology terms affecting biological processes that are regulated in heart failure are enriched among downregulated genes when using one-sided accumulative hypergeometric distribution analysis without multiple correction adjustment. c,d, Expression of genes that comprise ECM organization (c) and muscle contraction (d) gene ontology terms upregulated in TV-Dhet cardiomyocytes are normalized to CTRL cardiomyocytes (white bar). e, VCL expression levels are decreased in TV-Dhet cardiomyocytes compared to CTRL cardiomyocytes. DEseq2-adjusted P values displayed for TV-Dhet gene expression relative to CTRL cardiomyocytes. Gene expression P values were calculated using the Wald test with a Benjamini–Hochberg procedure false discovery rate adjustment. ****P < 0.0001. f, STRING database analysis reveals protein–protein interactions between the proteins encoded by upregulated genes, TPM1, VCL and limited high-confidence interactors. TPM1 and VCL are predicted to interact with upregulated components of the sarcomere (red oval) and costamere–ECM complex (blue oval), respectively. The darker edges represent increased STRING database interaction confidence.