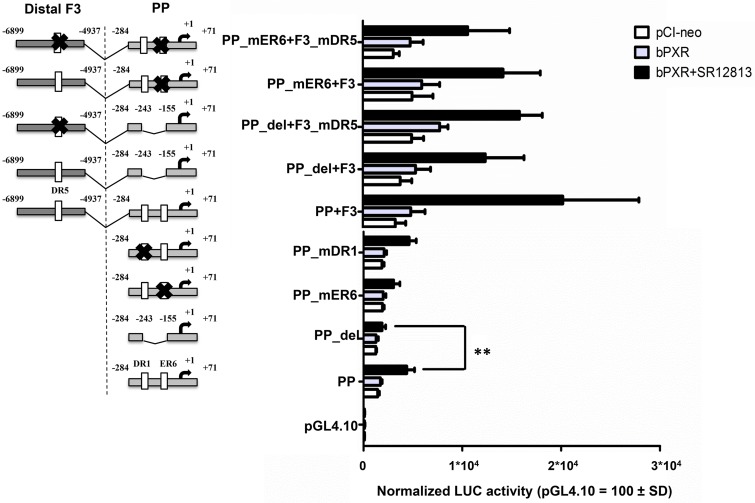
Fig 5. Identification of bPXR-responsive elements in the proximal promoter and fragment 3 in CYP3A28 promoter.
Several constructs were produced to study the binding elements identified in the CYP3A28 proximal promoter (PP) and the contribution of the binding motif DR5 identified in F3. The parental PP was deleted of the whole putative region containing several TF binding-sites resulting in the PP_del; through site-directed mutagenesis the ER6 (PP_mER6) and DR1 (PP_mDR1) motifs were inactivated. The parental PP+F3 was deleted of the whole putative region containing several TF binding-sites resulting in the PP_del+F3; through site-directed mutagenesis the ER6 (PP_mER6+F3), the DR5 motif (PP+F3_mDR5 and PP_del+F3_mDR5) or both (PP_mER6+F3_mDR5) were inactivated. Details are reported in S1 File. Numbers indicate the positions relative to the transcriptional start site. C3A cells were transfected with the control reporter pCMVβ (150 ng/well), each reporter plasmid or CYP3A4-XREM-luc (XREM, 50 ng/well) and either bPXR expression plasmid or pCI-neo empty vector (25 ng/well). After transfection, cells were treated with vehicle (0.1% DMSO) or SR12813 (10 μM) for 24 hours, and reporter activities were measured. Firefly luciferase activities were normalized with β-galactosidase activities. Data are expressed as relative activities to those in pGL4.10 transfected cells (= 100) for each condition (pCI-neo empty or bPXR co-transfection). Data are the mean ± SD (n = 3 or 4). Results shown are representative of 3 independent assays. Statistical significance P < 0.01: **, PP with ligand-treated bPXR vs. PP_del with ligand-treated bPXR.