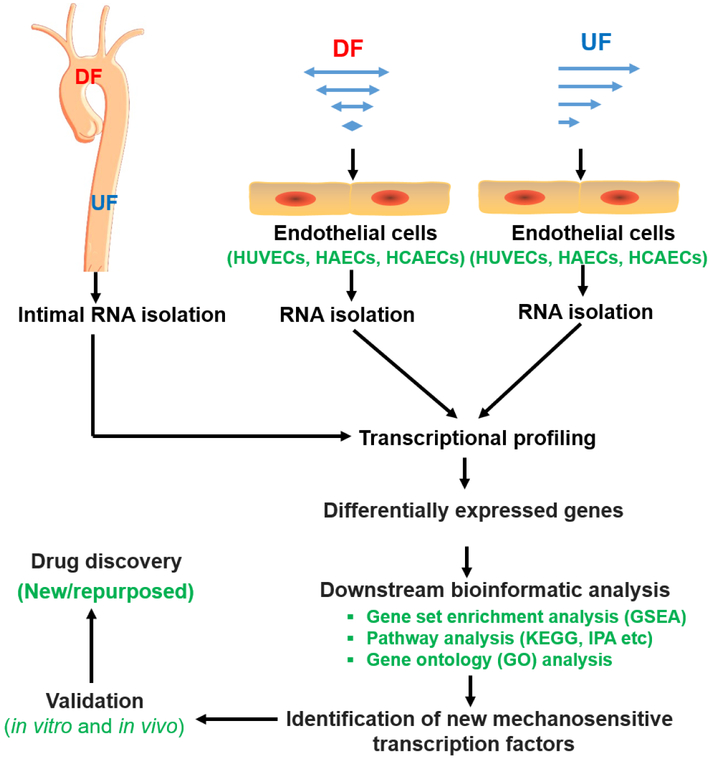
Figure 4. Discovery pipeline of novel MSTFs and targeted therapeutics.
Transcriptional profiling (such as RNA-sequencing or microarray) is commonly used to identify novel MSTFs. In vivo, intimal RNA samples from regions of DF and regions of unidirectional UF are collected for RNA-sequencing or microarray. In vitro, human ECs are exposed to UF, or DF before RNA is collected for transcriptional profiling. New MSTFs or gene regulation pathways can be identified by downstream bioinformatics analysis of differentially expressed genes. The newly identified MSTFs need to be validated both in cultured cells and in pre-clinical animal models. After validation, drug-screening system can be established to identify novel/repurposed pharmacological modulators of MSTFs, which can be further tested pre-clinically and clinically.
Abbreviations: HAEC, human aortic endothelial cells; HCAECs, human coronary artery endothelial cells; HUVECs, human umbilical vein endothelial cells; IPA, ingenuity® pathway analysis; KEGG, Kyoto encyclopedia of genes and genomes