Figure 8.
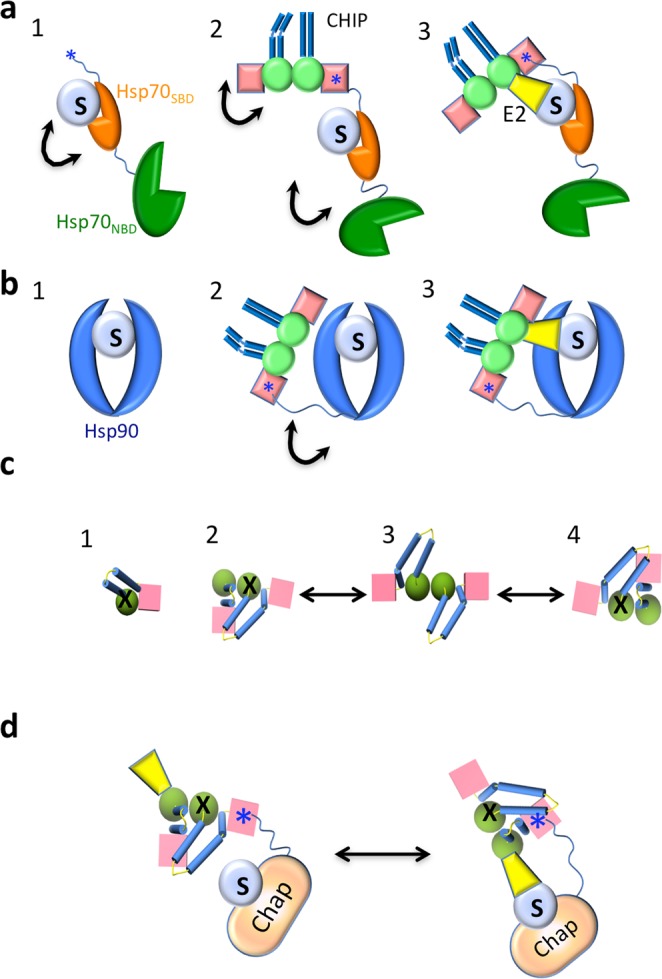
Flexibility in the interaction between chaperone, substrate and CHIP. (a) For Hsp70: (1) a flexible complex is formed through weak interactions between the Hsp70SBD and the substrate (S) (black arrows represent flexibility). (2) CHIP binds to Hsp70:S through the Hsp70 IEEVD motif (blue star) at the C-terminus of the chaperone and the TPR domain of one of the CHIP monomers. The EEVD motif is at the end of a long unstructured region (32 residues), which makes the Hsp70-CHIP interaction very flexible. (3) CHIP movements, together with binding of an E2 ubiquitin-conjugating enzyme (i.e., UbCH5a; yellow), allow substrate ubiquitination. (b) For Hsp90: (1) a flexible complex is formed through weak interactions between the Hsp90 substrate-binding region and the substrate. (2) CHIP binds to Hsp90:S through the MEEVD motif at the end of the long, unstructured region (32 residues) of one of the Hsp90 monomers and the TPR domain of one of the CHIP monomers, generating a flexible ternary complex with the substrate positioned between Hsp90 and CHIP. (3) CHIP movements, together with binding of an E2 enzyme, allow substrate ubiquitination. (c) Model of the conformational changes undergone by CHIP (structure observed from the top). The crystal structure of full-length CHIP is that of an asymmetric dimer7. (1) Each monomer is composed of a TPR domain (pink cube), a U-box domain (green sphere), and a dimerization domain (blue cylinders). Molecular dynamics studies50 have shown that the monomer in solution is stabilized through TPR-Ubox interactions which block the E2 binding site (X). (2) In the dimer, the two helix-coils come together such that one bends and generates two smaller, kinked helices. The dimer switches between the two possible asymmetric conformations (2 and 4) through a short-lived symmetric conformation (3). Dimer formation activates CHIP by unlocking one of the two E2 binding sites (the one without X). (d) Model of chaperone-bound, substrate ubiquitination mediated by CHIP and an E2 enzyme. When interacting with the chaperone:substrate complex, CHIP switches between the two asymmetric conformations described in (c), and E2 binding can alternate between the two potential E2-binding sites. Whereas the conformation on the left is that found in the 3D-reconstructed CHIP-based complexes in this study, the only productive interaction is that on the right, as it permits contact between the E2 enzyme and the substrate.
