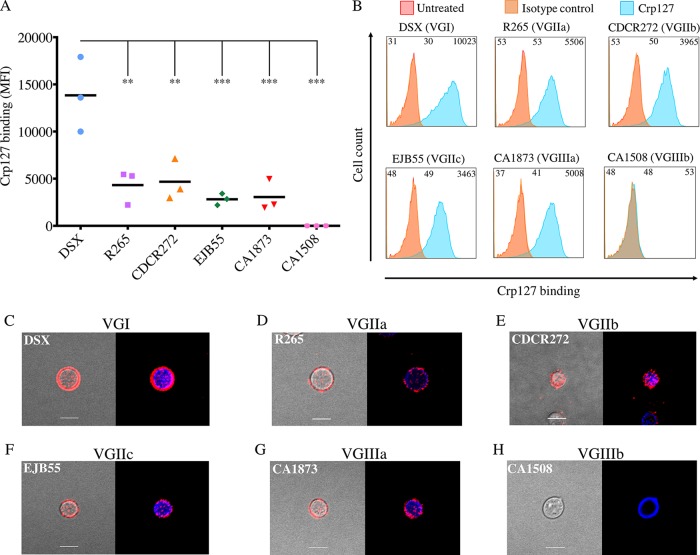
FIG 5.
Recognition of the Crp127 epitope is largely consistent within C. gattii serotypes. The ability of Crp127 to bind to six strains of C. gattii that encompass molecular types VGI to VGIIIb was quantified via flow cytometry. (A) Scatter plots showing corrected MFI values for each strain, which were calculated by subtracting the MFI value of isotype control cells from the MFI value of the corresponding Crp127-treated cells. Data points represent MFI values calculated from three biological replicates performed as independent experiments. Tukey’s multiple-comparison test was used to test the statistical significance of differences between the six strains (n = 3) (**, P < 0.01; ***, P < 0.001). (B) Histograms showing a representative distribution of Crp127 binding for strains DSX (type VGI), R265 (VGIIa), CDCR272 (VGIIb), EJB55 (VGIIc), CA1873 (VGIIIa), and CA1508 (VGIIIb). Fluorescence intensity values for untreated, isotype control, and Crp127-treated cells are presented, with the corresponding MFI values displayed at the top left, center, and right of each panel. (C to H) C. gattii strains DSX (C), R265 (D), CDCR272 (E), EJB55 (F), CA1873 (G), and CA1508 (H) were labeled for chitin (CFW) (blue) and Crp127 (goat Alexa 647-conjugated anti-mouse IgM μ-chain) (far red), and maximum-intensity projections were generated via confocal microscopy. Presented are representative images merged for transmitted light and Crp127 (left panels) and Crp127 and chitin (right panels). Bars, 5 μm.