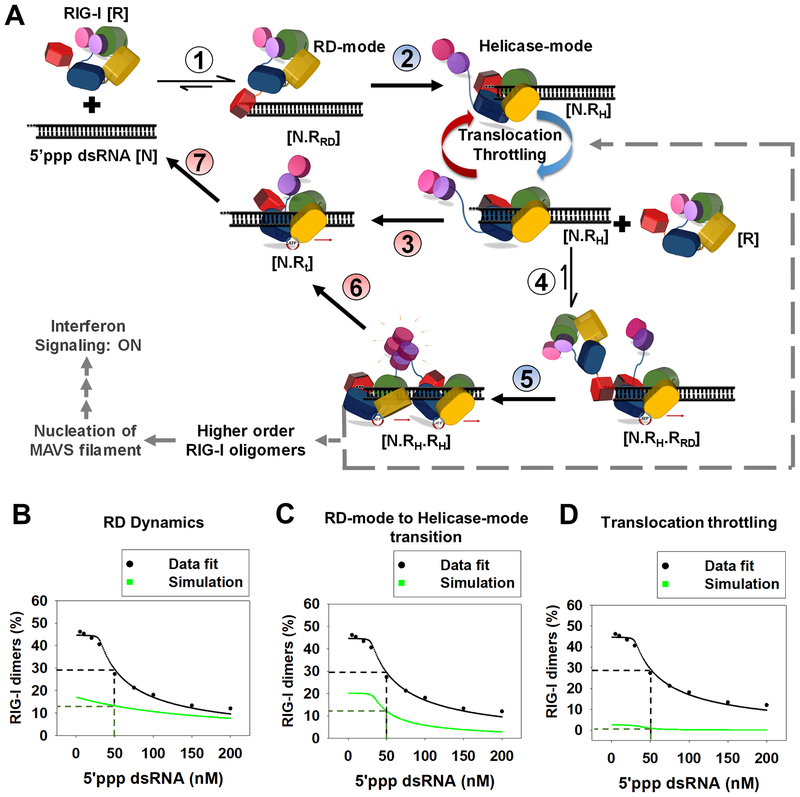
Figure 7. Kinetic proofreading pathway for RIG-I oligomerization and RNA discrimination.
(A) The seven steps show the kinetic pathway of RIG-I oligomerization. (1) RIG-I forms the RD-mode complex. (2) The RD-mode complex transitions into an ATPase active Helicase-mode complex that undergoes multiple rounds of ATP hydrolysis at the 5’-end (Translocation-throttling). (3) Helicase-mode complex translocates away from the dsRNA-end, or (4) binds a second molecule of RIG-I in the RD-mode. (5) Second RIG-I molecule transitions to Helicase-mode resulting in RIG-I dimer on dsRNA. (6–7) RIG-I dimer can translocate and dissociate from the RNA. Steps shaded in blue (2,5) are facilitated by ATP binding and steps shaded in red are driven by ATP hydrolysis (3,6 and 7). (B-D) The decrease in RIG-I dimers with increasing 5’ppp dsRNA concentration (Figure S6D) fit well to the kinetic proofreading pathway from A (black). The experimentally measured values used for the fitting are: Step 1 and 4 (kon: 6 × 108 M−1s−1, koff: 0.15 s−1), Step 3 and 6 (0.028 s−1), step 7 (1.2 s−1); predicted rates from best fit for step 2 and 5 were 0.03 s−1 and 0.06 s−1, respectively. Simulations (green) were carried out by either substituting the RD-mode off-rates of 5’ppp dsRNA in the model with 3’ovg dsRNA off-rate (B), decreasing the N.RRD to N.RH transition rate by 3-fold (C), and removing all translocation throttling steps at the 5’ end for monomer and dimer (D).