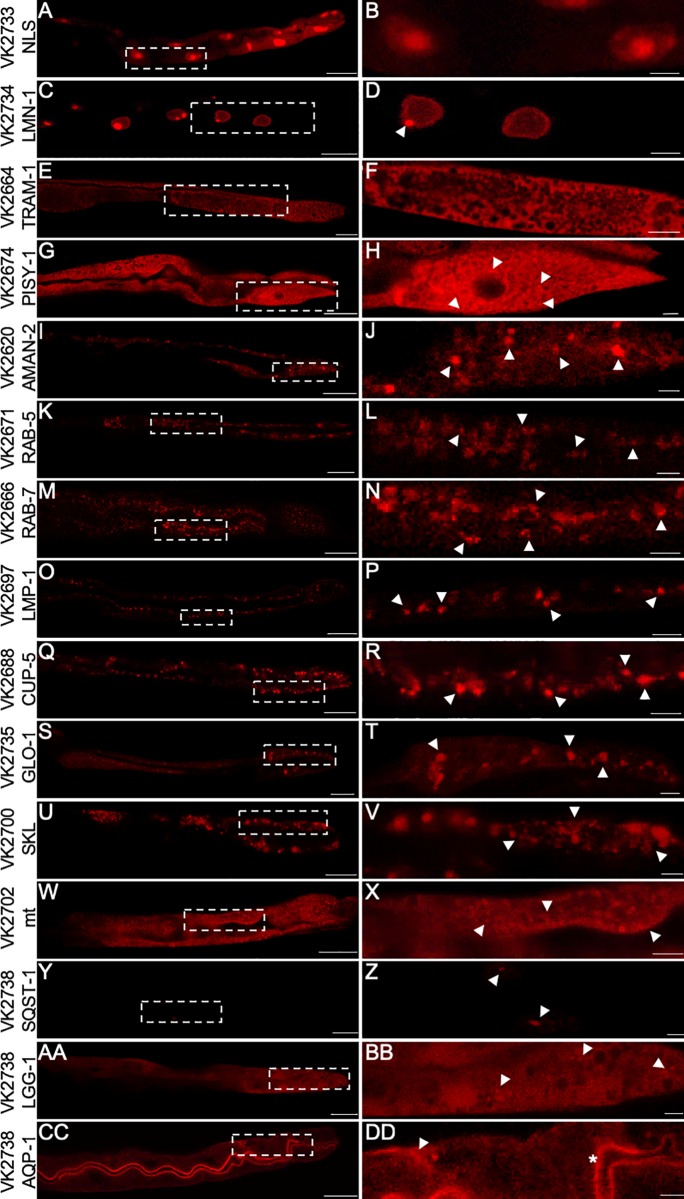
Fig 2. CemOrange2 subcellular expression patterns within C. elegans.
Representative confocal images of transgenic C. elegans strains expressing CemOrange2 fused with: the N-terminal SV-40 and C-terminal egl-13 nuclear localization signals (A, B); LMN-1, a nuclear lamin (C, D); TRAM-1, a rough ER protein (E, F); PISY-1, ER and Golgi protein (G, H); AMAN-2, a Golgi protein (I, J); RAB-5, an early endosomal protein (K, L); RAB-7, a late endosomal protein (M, N); LMP-1 and CUP-5, lysosomal membrane proteins (O-R); GLO-1, lysosomal related organelle protein (LRO) (S, T); SKL, a C-terminal peroxisomal targeting peptide (U, V); mt, an N-terminal mitochondrial target peptide (W, X); SQST-1 and LGG-1, autophagosome-related proteins (Y-BB); and AQP-1, an apical and basolateral plasma membrane protein (CC, DD). All transgenes were expressed using the intestinal specific promoter, Pnhx-2. Images were collected over >20 z-planes using a 40x PlanApo oil immersion objective (N.A. 1.3) at wavelengths Ex555 nm/Em565-590 nm. Either a maximum intensity projection (A—D) or a single XY-plane (E-DD) of the CemOrange2 expression are shown. The dashed box demarks a single intestinal cell to highlight the subcellular expression pattern shown in the right column (B, D, F, H, J, L, N, P, R, T, V, X, Z, BB, DD). Scale bars = 25 μm (left column) or 5 μm (right column). See text for description of arrowheads and asterisk.