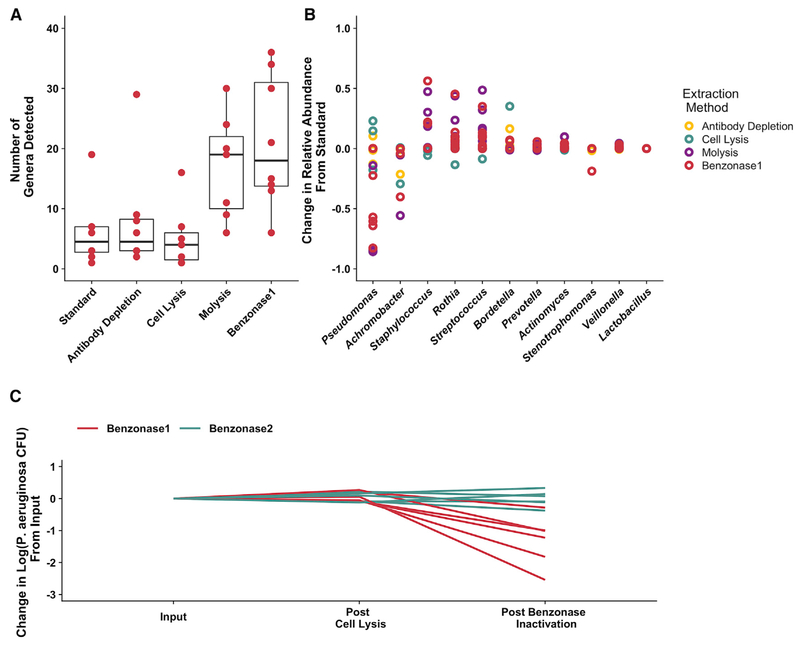
Figure 2. Effect of Extraction Method on Metagenomic Sequencing Taxonomic Profile and Viable Counts of the 8 Test Set 1 CF Sputum Samples.
(A) Number of distinct genera detected by metagenomic sequencing from DNA prepared with each extraction method. Results from extractions, including selective depletion of human and extracellular DNA, were compared to standard extraction conditions. Boxes represent the interquartile region, and black lines indicate the median value. Pairwise Wilcoxon signed rank tests were performed comparing each extraction method to standard extraction with a Benjamini-Hochberg correction for multiple comparisons, with no comparison reaching the level of significance (p < 0.05).
(B) Difference in relative abundance identified by each extraction method (compared with standard extraction) for the 10 most abundant taxa. Each data point indicates an individual sample.
(C) Effect of benzonase treatment on viable P. aeruginosa counts. Cultures of 6 separate P. aeruginosa clinical isolates were subjected to both benzonase1 and benzonase2 processing methods, which differ only in the order in which EDTA is added. Viable counts were measured after the hypotonic, host-cell lysis step and after enzyme inactivation, as indicated.
See also Figures S1–S4.