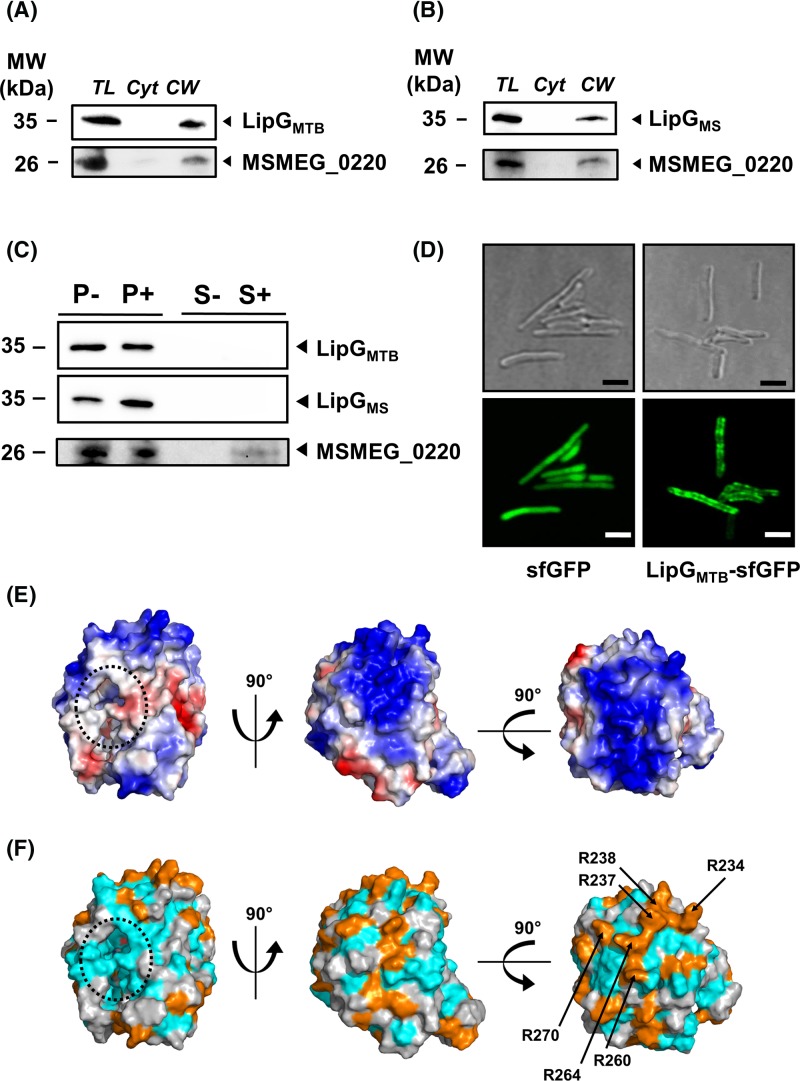
Figure 4. Subcellular location of LipG proteins.
(A,B) Subcellular localization of LipGMS and LipGMTB in M. smegmatis groEL1ΔC determined by ultracentrifugation. Recombinant cells expressing 6× His-tagged LipG were lysed, total lysate (TL), cytoplasm (Cyt), and cell wall (CW) fractions were separated by ultracentrifugation. Samples were loaded on to 12% SDS/PAGE and immunoblotted using HisProbe reagent. The exported MSMEG_0220 was used as control. (C) Determination of cell-surface-exposition of LipGMS and LipGMTB in M. smegmatis groEL1ΔC by detergent extraction. Recombinant cells expressing 6× His-tagged LipG were treated with PBS-buffer or with PBS-buffer containing Genapol®-X080 detergent. Pellet (P) and supernatant (S) fractions containing cytoplasmic and surface-exposed proteins respectively, were separated by centrifugation. Samples were loaded on to 12% SDS/PAGE and immunoblotted using HisProbe reagent. The exported MSMEG_0220 was used as a control. (D) Fluorescence microscopy analysis of M. smegmatis groEL1ΔC strains carrying either WT sfGFP gene or lipGMTB-sfGFP translational fusions. Cells were analyzed in both phase contrast (upper panel) and fluorescent channels (bottom panel). Scale bars represent 1 µm. (E) Electrostatic potential of LipGMTB 3D model. The electrostatic surface potentials were displayed color-coded on to a van der Waals surface using the PyMOL Molecular Graphics System (version 1.8, Schrödinger, LLC). Red and blue colors represent net negative and positive charges, while white color represents overall neutral positions, respectively Black lines are showing the catalytic pocket. Two rotations of 90° were performed in order to provide a better view of the high potential area on top of the protein. (F) Position of positively charged residues in LipGMTB 3D model. Positives residues were highlighted in orange. The two rotations of 90° were conserved in order to provide a better view of the positively charged residues on top of the protein. An arginine-rich patch has been identified and position of the respective residues (R234, R237, R238, R260, R264 and R270) are marked with black arrows.