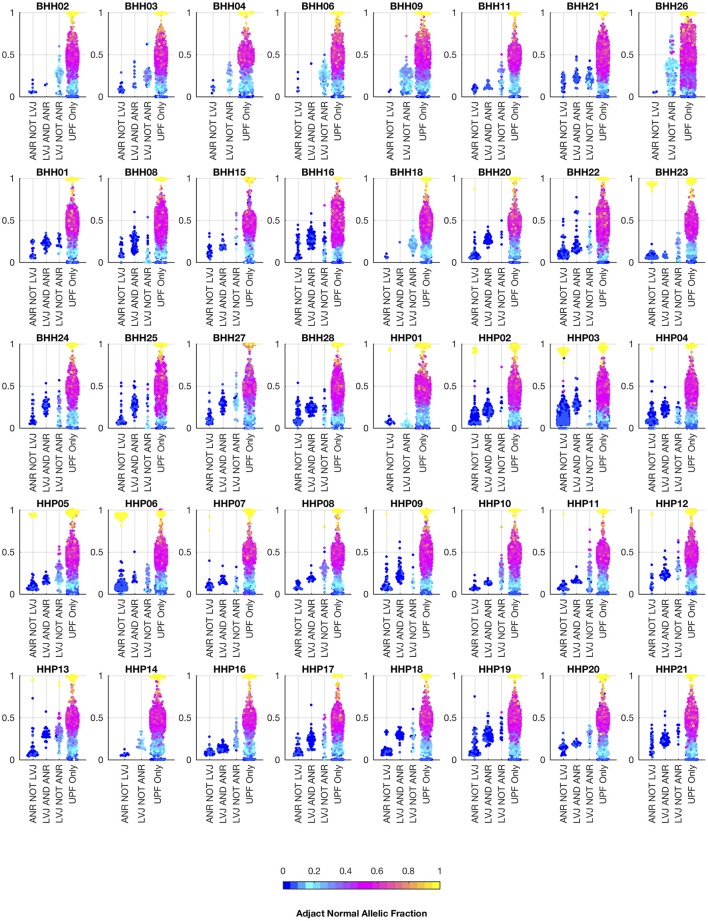
Figure 7.
Comparison of allelic fractions of variants in archival dataset by calling method. For each of the breast and prostate patients, the allele fractions in the tumor sample are plotted for the variants detected in each of the three approaches. The color of each point indicates the allele fraction of the variant in the adjacent normal sample. Most of the variants detected in the adjacent normal as reference approach, but not lumosVar 2.0 joint analysis (ANR NOT LVJ), have low allele fractions in both the tumor and the adjacent normal. The variants detected by lumosVar 2.0 joint analysis, but not adjacent normal as reference approach (LVJ NOT ANR) typically have higher allele fractions in the tumor, and lower allele fractions in the adjacent normal, though lumosVar 2.0 joint analysis also detects some variants that are lower allele fraction in the tumor and higher allele fraction in the adjacent normal in a few patients such as HPP01. The variants only called in the unmatched filtering (UPF only) approach have similar allele fractions in the tumor and adjacent normal samples. The 8 patients in the top row had the adjacent normal tissue macrodissected from tumor containing slides and these patients typically have more variants detected by lumosVar 2.0 joint analysis and not ANR compared to the remaining patients whose adjacent normal sample was procured from separate slides.