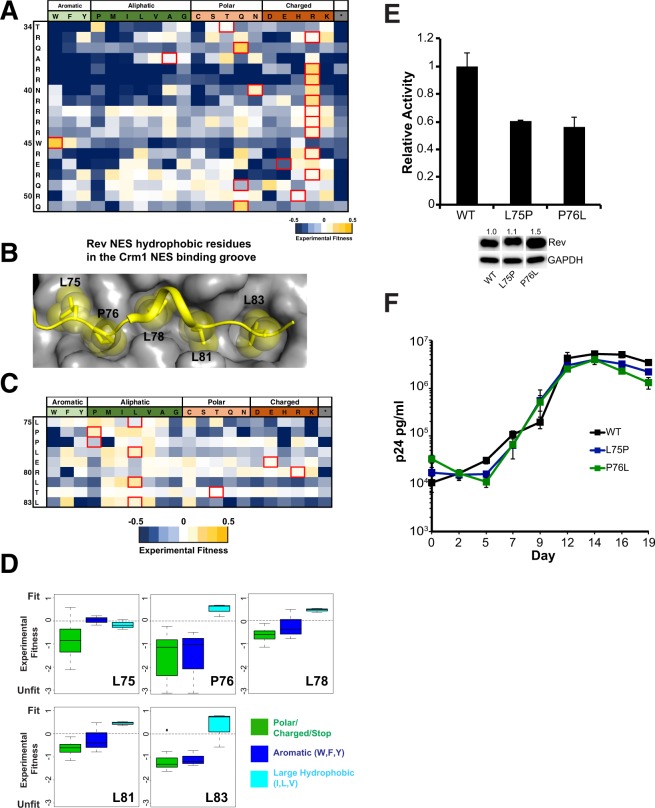
Figure 3.
Mutational limits of the Rev ARM and NES. (A) CDMS fitness values for residues in the Rev ARM10 (B) Structure of Rev NES (yellow) bound to Crm1 (grey) (PDBID: 3NBZ) with the Rev hydrophobic residues binding to the Crm1 NES binding groove shown as spheres (C) CDMS fitness values for the NES10 (D) Preferences for Rev NES contact residues using CDMS fitness values10. (E) Reporter assay probing export activity of Rev NES mutations. Data are mean ± standard deviation (s.d) of biological replicates. Western blots below show protein expression for Strep-tagged Rev and GAPDH as loading control, and numbers above blots represent quantified band intensities for Rev mutants relative to WT-Rev. The original blot for the cropped bands is shown in Fig. S5. (F) Viral replication profiles of viruses containing Rev with NES mutations. Data are mean ± s.d of biological triplicates.