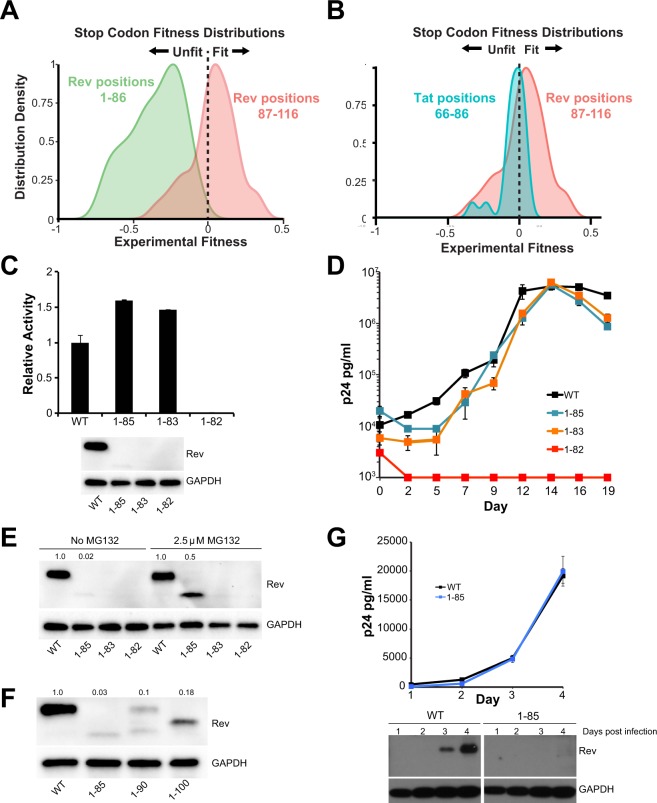
Figure 5.
The HIV-1 Rev C-terminus is not required for function but regulates Rev stability. (A) Stop codon fitness distributions from CDMS dataset for Rev10. (B) Stop codon fitness distributions from CDMS dataset for Tat C-terminus (blue, residues 66–86) and Rev C-terminus (red, residues 86–116) (stop codon data for both proteins10. (C) Reporter assay probing export activity of Rev C-terminal truncations. Data are mean ± standard deviation (s.d) of biological replicates. Western blots below show protein expression for Strep-tagged Rev and GAPDH as loading control. (D) Corresponding viral replication spread experiments. Viral p24 levels below 1000 pg/ml are shown as 1000 pg/ml in the plots for illustration purposes. Data are mean ± s.d of biological triplicates. (E) Adding MG132, a proteasome inhibitor partially restores expression of Rev with C-terminal truncations in transiently transfected 293 T cells. Numbers above blots represent quantified band intensities for Rev mutants relative to WT-Rev. (F) Shortening the length of C-terminal truncations does not restore protein expression transiently transfected 293 T cells. Here, we note that we consistently observe two bands for Rev 1–90, one with a faster mobility and the second with the same mobility as full-length Rev. Numbers above blots represent quantified band intensities for Rev mutants relative to WT-Rev. (G) C-terminal truncations affect Rev expression/stability from HIV-1 (with Flag-tagged Rev in the nef locus) in infected T-cells. Plot shows virus production from cell supernatant, quantified by p24 ELISA. Western blots below show protein expression for Flag-tagged Rev and GAPDH loading control from the cells. The original blot for the cropped bands is shown in Fig. S5. Data are mean ± s.d of biological replicates.