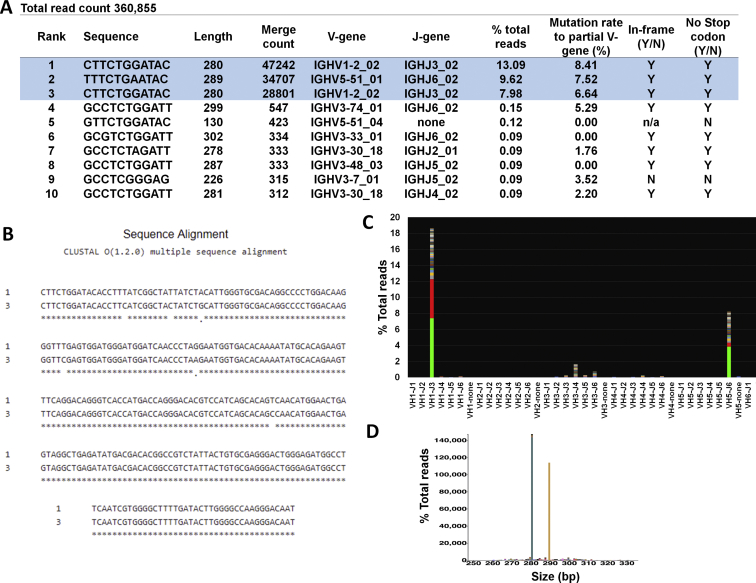
Supplemental Figure S4.
Representative example requiring additional merging in the final analysis. A: On the basis of the merged read summary output, there are three dominant sequences (highlighted in blue). Sequences ranked 1 and 3 have the same length and VJ use. B: When aligned, sequences are related, differing by six bases, C to T and G to A substitutions typical of somatic hypermutation events. After additional merging, there are two separate clones: clone 1: 280 bp, V1-2-02;J3, 21.07% of total reads (13.09 + 7.98); and clone 2: 289 bp, V5-51_01;J6, 9.62% of total reads. The two sequences are 140× and 64× of the background, respectively (21.07/0.15 and 9.62/0.15, respectively). In this case, the background starts at the fourth sequence. Both clones meet criteria for clonality. C: Snapshot of the V-J sequence frequency graph of LymphoTrack MiSeq Software version 2.3.1 shows two clonal sequences with same use (VH1-J3) stacked and the third sequence at V5;J6. D: Sequences plotted by the Memorial Sloan Kettering LymphoClone viewer show sequences merged by VJ family use and size. Correlation with corresponding flow cytometry shows the bone marrow sample is involved by both chronic lymphocytic leukemia and plasma cell neoplasm. N, no; n/a, not applicable; Y, yes.